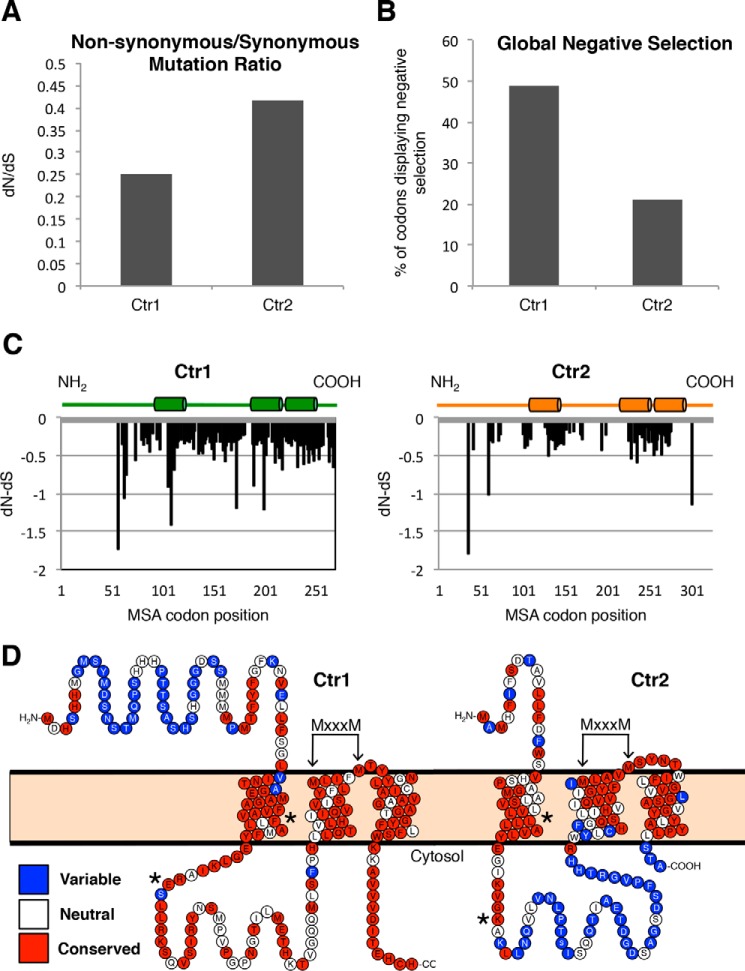
Figure 2.
Ctr2 demonstrates relaxed purifying selection. A, predicted Ctr family genes from 51 metazoan species were aligned, and single-likelihood ancestor counting analysis was performed under standard parameters using the Datamonkey server. Global dN/dS values are reported. B, the number of individual codons from the Ctr1 or Ctr2 DNA alignment that display negative selection (p ≤ 0.05) were divided by the number of total codons in the Ctr1 or Ctr2 alignment, respectively. C, normalized dN/dS values for each codon displaying selection (p ≤ 0.05) for Ctr1 and Ctr2 alignments are plotted. Cylinders represent transmembrane domains. D, protein sequences from metazoan Ctr1 and Ctr2 were aligned and analyzed for conservation with the ConSurf server using a Bayesian evolution method. Shown are the human Ctr1 and Ctr2 proteins with amino acids that represent variable (blue), neutral (white), or conserved (red) positions. Arrows point to the highly conserved MXXXM motif necessary for copper transport. Asterisks denote residues Phe-77 and Glu-91 in human Ctr1 as well as Leu-34 and Lys-47 in human Ctr2.