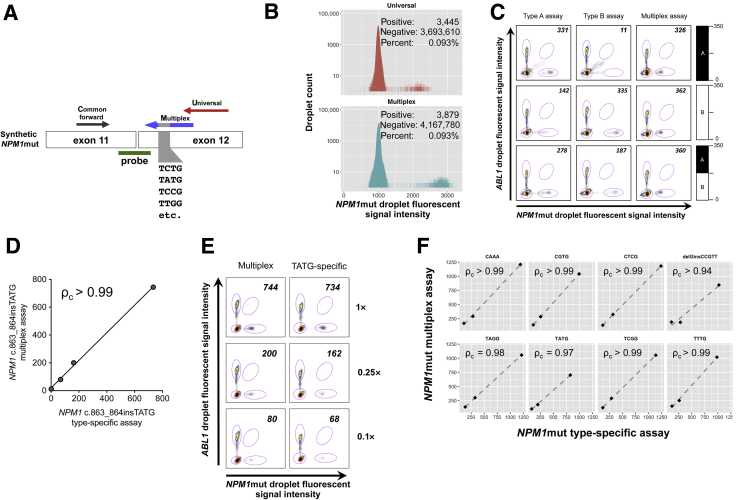
Figure 3.
Multiplex NPM1 assay accurately detects rare NPM1 mutation (NPM1mut) types. A: Schematic representation of synthetic NPM1mut target consisting of a pool of degenerate NPM1mut insertion sequences. Amplification is performed using common forward primer with multiplex reverse primer pool (mutant specific) or the universal reverse primer (amplifies both mutant and wild-type). Common probe is used for all reactions. B: Histogram indicating counts and distribution of fluorescence signal intensity of NPM1-positive and -negative droplets. C: Plasmids harboring type A (top row), type B (middle row), or a 50:50 mixture of type A and B (bottom row) NPM1 mutations were spiked into GM12878 background cDNA. A target of 350 NPM1mut/104ABL1 copies was used in all cases and the expected content indicated graphically by the bar plot on each row. Detection was attempted by digital PCR (dPCR) using the specific assays for the type A or type B mutation alongside the massively multiplex assay. The vertical axis of each dot plot indicates the signal intensity of ABL1-positive droplets, and the horizontal axis indicates NPM1mut-positive droplets. NPM1mut/104ABL1 ratios are indicated in the upper right corner. D: Synthetic template for NPM1 c.863_864insTATG is spiked into GM12878 cDNA targeting approximately 1000 NPM1mut/104ABL1 copies and left undiluted (1×) or diluted fourfold (0.25×) or 10-fold (0.1×) alongside negative control cell line GM12878. The axes indicate NPM1mut/104ABL1 as determined by the NPM1 multiplex assay (vertical axis) versus type-specific assay (horizontal axis). E: Scatterplot of dPCR data for the multiplex assay and c.863_864insTATG-specific assay adapted to dPCR for the dilution series from A with dilutions 1×, 0.25×, and 0.1× shown. F: Correlation between NPM1mut/104ABL1 ratios for a different synthetic rare NPM1 insertion mutations spiked into GM12878 cDNA targeting an approximately 1000 NPM1mut/104ABL1 starting ratio. Undiluted spike-in mixture (1×) is then compared with fourfold (0.25×) and 10-fold (0.1×). The position 863 insertion sequence is indicated over each plot except for delGinsCCGTT, which is NPM1 c.864_865delGinsCCGTT. The axes indicate NPM1mut/104ABL1 ratios for the multiplex assay (vertical axis) versus type-specific assays (horizontal axis). Lin's concordance correlation coefficient is indicated (ρc).