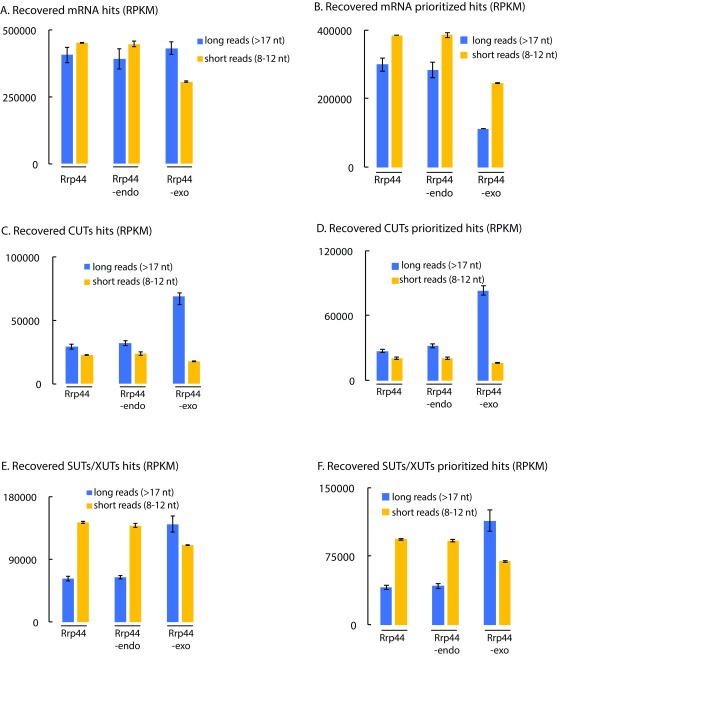
Figure 4. Short reads are preferentially mapped to mRNAs, SUTs and XUTs compared to CUTs.
( A– F) RPKMs were calculated for Rrp44, Rrp44-endo and Rrp44-exo and summed for all mRNAs ( A, B), CUTs ( C, D) and SUTs/XUTs ( E, F) using default counting of overlaps with genes ( A, C, D) or prioritized counting ( B, D, F). RPKM for long (blue) and short (yellow) reads are averaged between two or three independent experiments and shown with standard deviation.