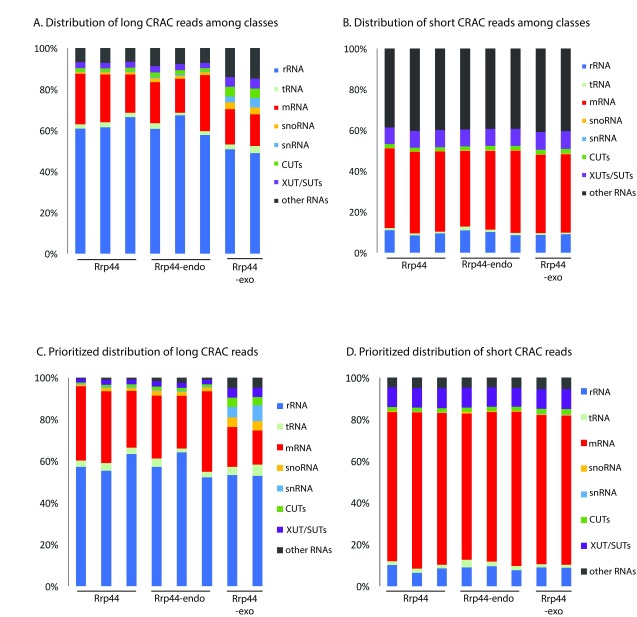
Figure 2. Mapping of long and short reads among RNA classes.
( A– D) Distributions of long ( A, C) and short ( B, D) mapped reads recovered in CRAC datasets between RNA classes, using default counting of overlaps with genes output ( A, B) or prioritized count ( C, D). For prioritized alignment, RPKM values were calculated for each long read aligned to the genome, sorted by value and then used as priority order for reads aligning to different places in the genome, to reduce mis-mapping (see Materials and Methods). Two or three biological replicates are shown for each protein.