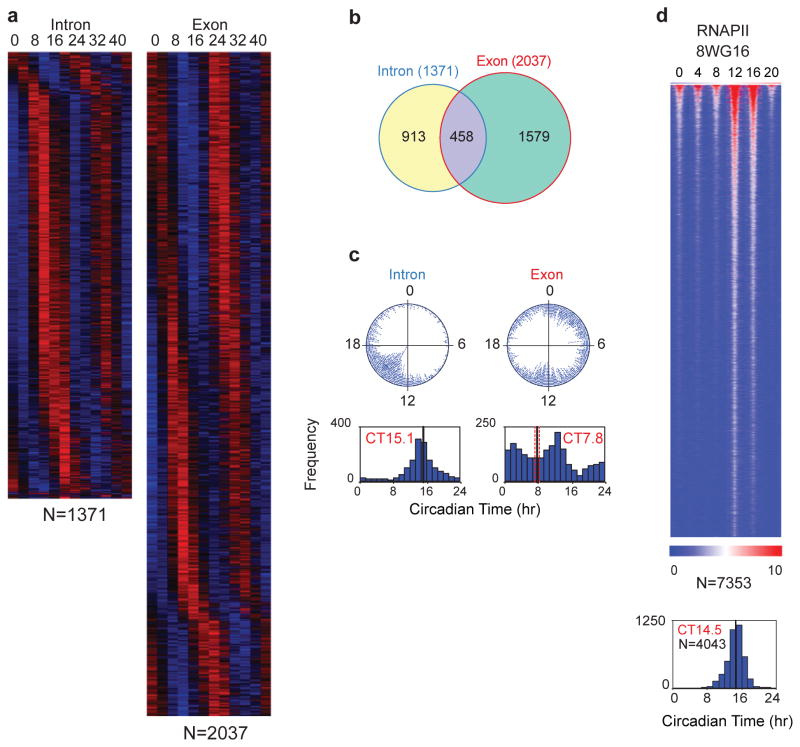
Figure 4.
Whole-transcriptome RNA-seq analysis of circadian gene expression in the mouse liver. a) Heat map view of intron (left) and exon (right) RNA cycling genes. Each gene is represented as a horizontal line, ordered vertically by phase in hours indicated at the top. Twelve time points are shown beginning at CT0 and ending at CT44 from left to right. “N” indicates the number of cycling genes in each category. b) Venn diagram of intron and exon cycling genes showing only 22% overlap. c) The phase distribution of cycling genes. The phase of each transcript rhythm is represented in a dot plot (top) and histogram plot (bottom). d) Heat map view of cycling RNAPII-8WG16 occupancy across the genome of the liver with a peak occurring at night. Modified/Reproduced with permission from REF.71.