Figure 2.
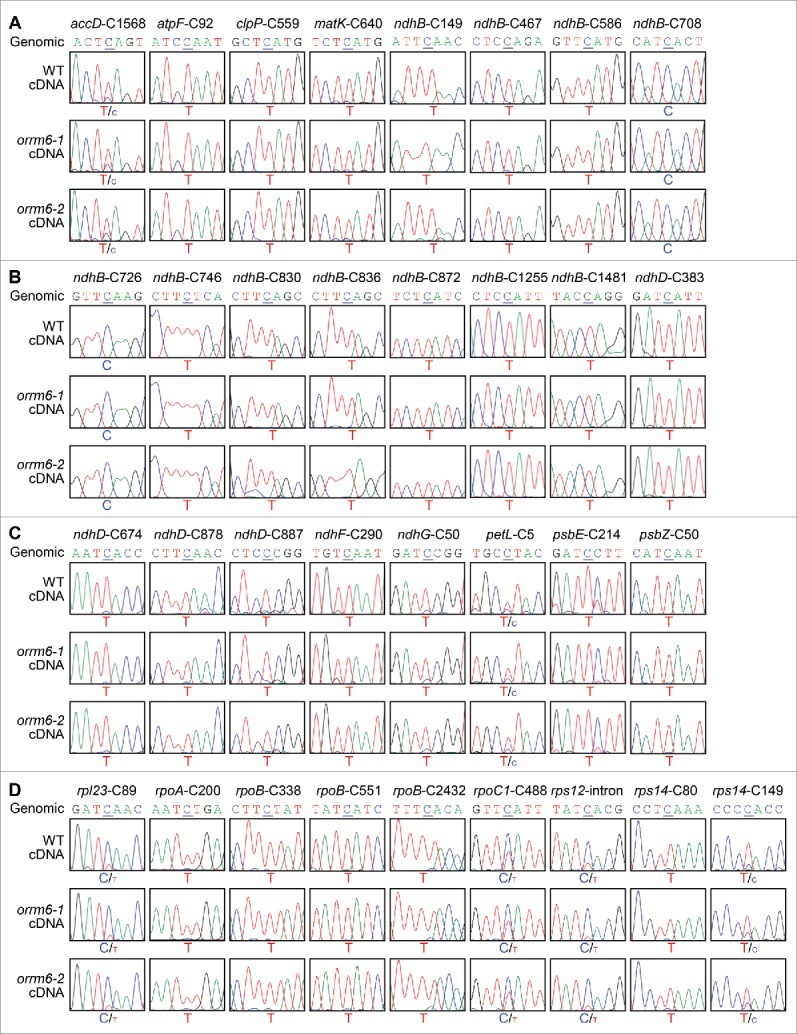
Sanger sequencing of plastid RNA editing sites in the wild-type and orrm6 mutant plants. (A) Analysis of RNA editing at accD-C1568, atpF-C92, clpP-C559, matK-C640, ndhB-C149, ndhB-C467, ndhB-C586, and ndhB-C708. (B) Analysis of RNA editing at ndhB-C726, ndhB-C746, ndhB-C830, ndhB-C836, ndhB-C872, ndhB-C1255, ndhB-C1481, and ndhD-C383. (C) Analysis of RNA editing at ndhD-C674, ndhD-C878, ndhD-C887, ndhF-C290, ndhG-C50, petL-C5, psbE-C214, and psbZ-C50. (D) Analysis of RNA editing at rpl23-C89, rpoA-C200, rpoB-C338, rpoB-C551, rpoB-C2432, rpoC1-C488, rps12-intron, rps14-C80, and rps14-C149. RT-PCR products surrounding the editing sites were directly sequenced. The 7-nucleotide sequences encompassing the cytidine target (underlined) were shown. The corresponding genomic sequences of these 2 sites were displayed as controls.
