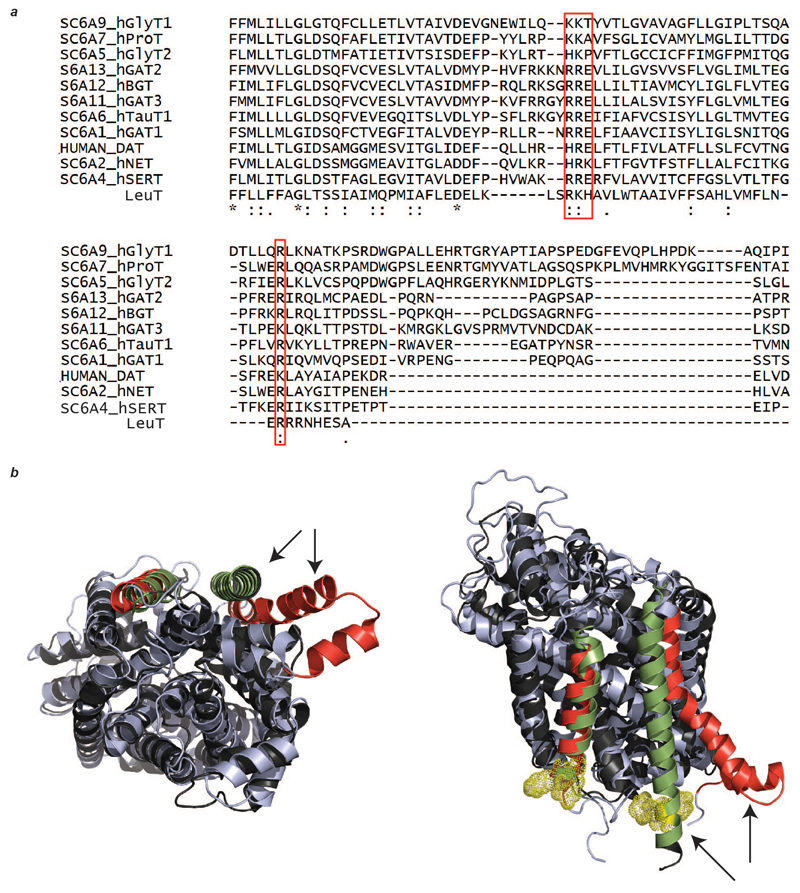
Extended Data Figure 8. Sequence and structure alignment of LeuT with other eukaryotic biogenic transporters.
a, The basic residues of LeuT that are involved in lipid binding (red box) are conserved across the BATs. b, Two views of the superimposed structures of LeuT (PDB ID 2A65, black) and SERT (PDB ID 5I6Z, light blue) show the differences in the dimer interface. Dimer interface helices are highlighted with arrows and coloured (LeuT green, SERT red); basic residues responsible for lipid binding in LeuT (yellow mesh). One of the interface helices in SERT swings away from the interface, negating the possibility of lipid-induced oligomerisation analogous to that proposed for LeuT.