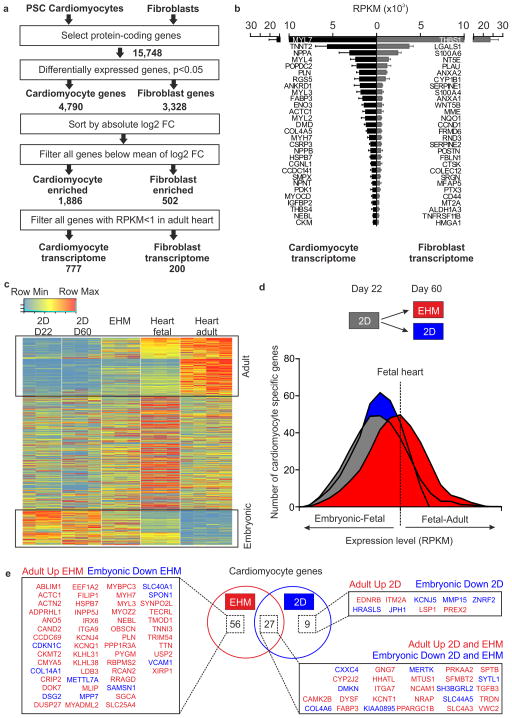
Figure 3. Molecular maturation of serum-free EHM.
(a) Strategy to determine cardiomyocyte and fibroblast transcriptomes from RNAseq data obtained from purified pluripotent stem cell-derived (PSC) cardiomyocytes (n=3 hES2 RFP, n=3 iCell CM, n=3 hiPS-G1) and primary fibroblasts (n=3 HFF, n=3 human cardiac fibroblasts, n=3 human gingiva fibroblasts). (b) RPKM values of the 29 most abundantly expressed transcripts in PSC-derived cardiomyocyte and primary fibroblasts. (c) Heatmap of cardiomyocyte transcripts in 22 day old cardiomyocyte monolayer cultures (2D D22), 60 day old cardiomyocyte monolayer cultures (2D D60), 6 week old EHM (note that cardiomyocyte “age” in these EHM was similar to 2D D60 cultures), fetal heart, and adult heart. Boxed areas indicate cardiomyocyte maturation genes; “adult”: increasing expression with development (upper box), and “embryonic”: decreasing expression with development (lower box). (d) Histogram of cardiomyocyte gene expression level (RPKM) compared to fetal heart as reference. Comparison of 22 day old cardiomyocyte monolayer cultures (2D, grey box) as starting point, 60 day old cardiomyocyte monolayer cultures (2D, blue box), and 6 week EHM cultures (red box). (e) Venn diagram and corresponding list of differentially expressed cardiomyocyte maturation genes with specific regulation in EHM, 60 day old cardiomyocyte monolayer cultures (2D), or both (overlap in Venn diagram; p<0.05 corrected for multiple testing by Benjamini-Hochberg method).