Figure 1.
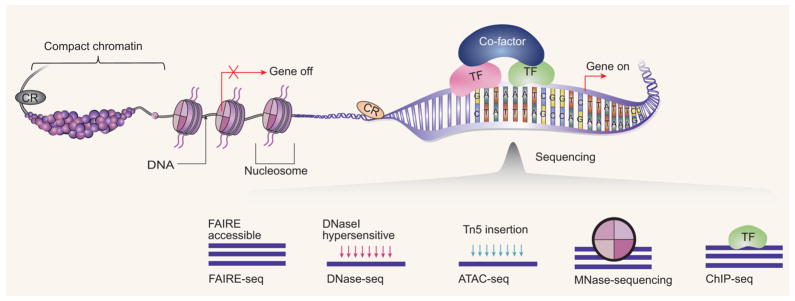
The basic concepts for open chromatin. Chromosomal DNA is compacted inside a nucleus by hierarchically folding DNA into certain chromatin structures. Most of the DNA fragments are compact chromatin and are tightly wrapped around histones (the cylinders are the core histones and around which the DNA is wrapped). Open chromatin are the DNA fractions which are accessible to the transcriptional machinery (including the bound TF and cofactors to regulate genes and the chromatin regulator (CR) modulating the chromatin state.) and further influences gene expression (turn the genes on or off). The rapidly developing sequencing methods, such as FAIRE-seq, DNase-seq, ATAC-seq, MNase-seq, and ChIP-seq, together provide the necessary information to decode the regulatory landscape inside cell. Those techniques utilize different mechanisms and provide complementary information. The FAIRE assay enriches for such open chromatin regions by differential solubility in phenol. The DNase I assay utilizes the fact that regions of the open chromatin are much more susceptible to DNase I digestion. ATAC assay integrates sequencing adaptors into regions of accessible chromatin by Tn5 transposase. MNase assay uses micrococcal nuclease to digest chromatin to study nucleosomes. The ChIP assay uses a specific antibody to enrich for DNA regions binding to a specific TF or a modified histone. Modeling the massive data generated by those technologies allows us to reveal the interplay among TF binding, active TSS, nucleosomes and nucleosome modifications, enhancers, and insulators in a wide variety of cell lines and tissue samples. Particularly causal regulatory network inference is promising by integrating the information from chromatin level with the gene expression data.