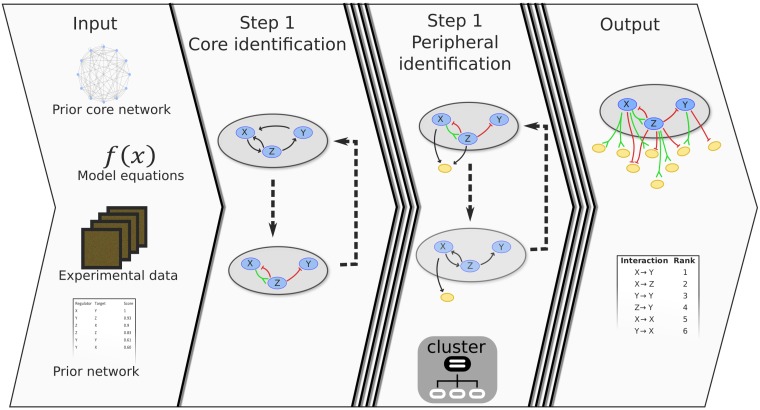
Fig 1. General workflow of the LArge-Scale SIMulation (LASSIM) high performance toolbox.
As Input LASSIM take four basic components, core and peripheral prior networks (optional), experimental data and dynamic equations to fit a large-scale non-linear dynamic system based on the fully parallel PyGMO toolbox. Step 1, LASSIM performs pruning and data fitting on the core system. Step 2, LASSIM expands the core model by inferring outgoing interactions with peripheral genes, with each gene solved in parallel using a computer cluster. The LASSIM functions are fully modular, and have been built so that the functions describing the optimization procedure, dynamic equations, cost function and data pruning are modular and can easily be changed by the user. The Output is either the core network defined in Step 1 or a genome wide regulatory network if Step 2 is run, with ranked interactions based on selection order, as well as kinetic parameters for the dynamic equations.