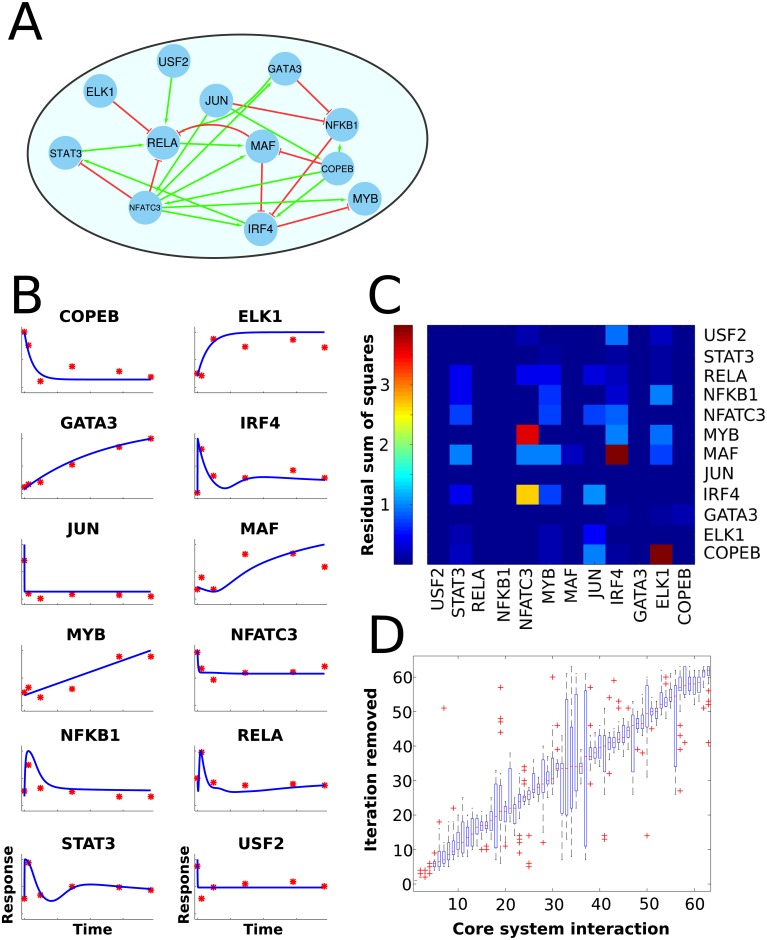
Fig 2. LASSIM inferred a robust minimal and full-scale non-linear transcription factor—Target dynamic system describing naïve T-cells towards Th2 cells.
(A) We identified 12 core Th2 driving TFs from the literature, and inferred their putative targets using DNase-seq data from ENCODE. In total, these interactions constituted of 63 core TF-TF interactions and 64,872 core-to-peripheral gene regulations. These interactions were assumed to follow a sigmoid function, as described in the Methods section. The complete prior network was, together with Th2 differentiation dynamics and siRNA mediated knock down data of each TF measured by microarray profiling, used by LASSIM to infer a Th2 core system. As can be seen in the core network, there are feedback loops between several of the TFs. (B) Microarray time series experiments (red dots) and respective state simulated by the LASSIM model (blue solid lines) of the core TFs. On the x-axis is time, and the y-axis denotes gene expression in arbitrary units. (C) Heat map of the data fit of the Th2 model to the siRNA perturbation data, i.e. the siRNA part of V(p*). Each siRNA knock-down experiment is represented as a separate column. For example, the model fits the response of a siRNA knock-down on IRF4 well for all TFs except MAF well. (D) Box-plot representing the ranking of each removed parameter from multiple stochastic optimizations of the core model. All edges that had a median selection rank over 40 were included in the final model. Model selection was based on prediction error variation, see section model selection and S3 Fig.