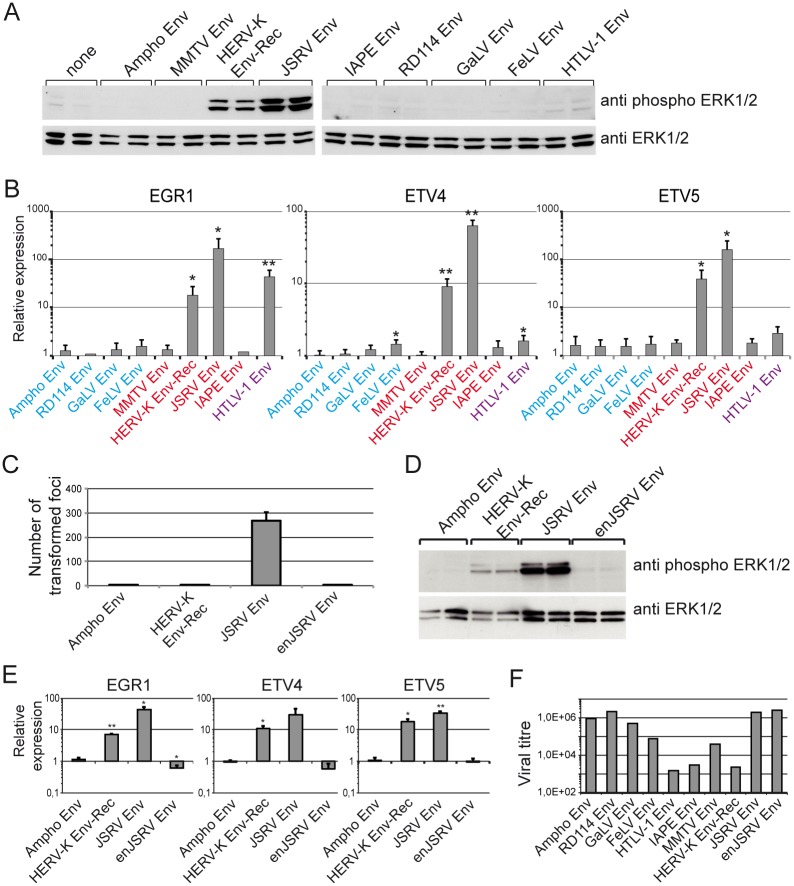
Fig 4. Oncogenic properties carried by different retroviral Envs.
(A) Phosphorylation of ERK1/2 in 293T cells expressing a panel of retroviral Envs was assayed as described in Fig 3B. Samples in left and right panels were prepared and processed (migration, transfer, revelation) at the same time; therefore intensity of the signals in both sets of panels can be compared directly. The Envs tested span three different retroviral genera and are colour-coded according to group (red: betaretroviruses; blue: gammaretrovirus; purple: deltaretrovirus). (B) The mRNA levels of the transcription factors EGR1, ETV4 and ETV5 following expression of the indicated Envs were measured as described in Fig 2C. Error bars represent standard deviation of the mean of three independent experiments. (C) The transforming activity of Ampho, HERV-K, JSRV and enJSRV retroviral Env proteins was assessed in 208F cells. Briefly, cells were transiently transfected with expression vectors for the 4 Env proteins and left to reach confluence. The number of transformed foci was counted after 3–4 weeks. The histogram represents the average number of foci obtained per 2x105 cells in 3 independent experiments. (D) Phosphorylation of ERK1/2 in 293T cells expressing the 4 retroviral Envs mentioned above was assessed as in Fig 3B. (E) The mRNA levels of transfection factors EGR1, ETV4 and ETV5 following expression of the 4 Envs was tested as described in Fig 2C. (F) We ensured that all the expression vectors for the Env proteins were functional by measuring the viral titres obtained with them when pseudotyping heterologous retroviral particles. Ampho, RD114, GaLV, FelV and MMTV Envs were tested using a MLV core, HERV-K, JSRV, enJSRV, IAP and HTLV1 Envs were tested with a HIV core. Viral titres were measured on 293T cells, except for FeLV pseudotypes (feline G355.5 cells) and MMTV pseudotypes (mouse NIH 3T3 cells). Background infection levels (i.e. the titres obtained with no Env expression vector) were less than 100 whatever the core/target cells combination.