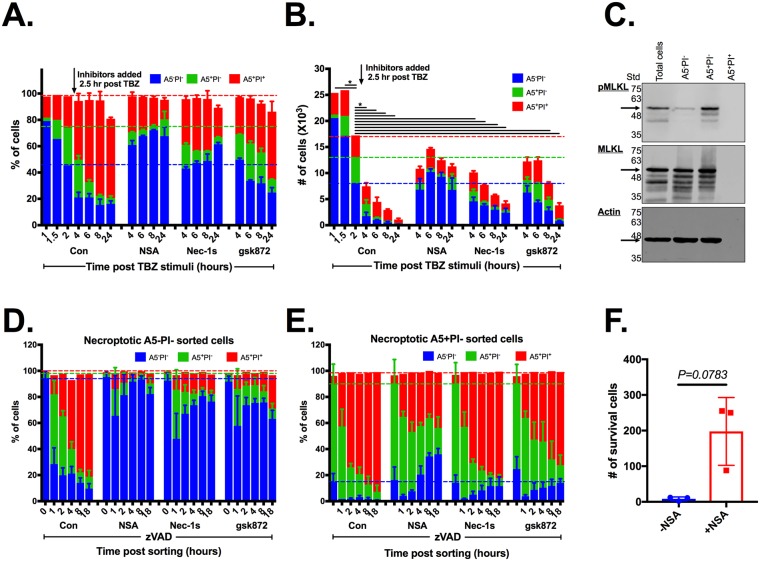
Fig 5. Necroptotic A5 single-positive cell death requires pMLKL membrane translocation.
U937 cells were stimulated for necroptosis using TNFα, birinapant (SMAC mimetic) and zVAD (TBZ). After 2 hours, cells were treated with necroptotic inhibitors RIPK1 (Nec-1s), RIPK3 (gsk872), and pMLKL (NSA), or left untreated (Con). (A) Cell viability was measured at different time points after stimulation of cell death using A5/PI staining and then analyzed by flow cytometry (mean ± SD). (B) Cell counts were measured using the Attune NxT flow cytometer (mean ± SD). Statistic comparisons between total cell numbers in each group to total cell numbers at the 2 hour time point in the control group was carried using ANOVA, followed by a Tukey’s multiple comparison test, * p < 0.05. Data are representative of 1 experiment from at least 3 independent ones. (C–F) U937 cells were stimulated as in (A) for 2 hours, prior to A5/PI staining. Cells were sorted into live cells (double-negative) or PS-exposed-necroptotic (A5-positive PI-negative). (C) Necroptotic cell death key factor pMLKL was detected in the different sorted populations using western blot. Cell viability was measured at different time points after sorting in the A5−PI− (D) and A5+PI− (E) populations, using A5/PI staining, and then analyzed by flow cytometry (mean ± SD). Where indicated, RIPK1 (Nec-1s), RIPK3 (gsk872), and pMLKL (NSA) inhibitors were added to the collection tubes. Dashed lines represent the state of cells prior to the addition of inhibitors. Data are taken from 3 independent experiments. (E) U937 cells were stimulated and sorted into PS-exposed-necroptotic (A5-positive PI-negative) as above. Viable cells (A5−PI−) were measured and counted at 6 days after sorting. Data are taken from 2 independent experiments. (mean ± SD). Statistic comparisons between total cell numbers was carried using parametric paired Student t test. All raw data for the data summarized under this Fig can be found in S5 Data. A5, annexin V; Con, control; Nec-1s; necroptotic inhibitor of RIPK1; MLKL, mixed lineage kinase domain-like; NSA; necrosulfonamide; PI; propidium iodide; pMLKL; phosphorylated mixed lineage kinase-like protein; PS, phosphatidylserine; RIPK1, receptor-interacting protein kinase-1; RIPK3, receptor-interacting protein kinase-3; SD, standard deviation; SMAC, second mitochondrial-derived activator of caspase; Std, protein ladder; TBZ, TNFα + birinapant + zVAD; zVAD, Z-VAD-FMK.