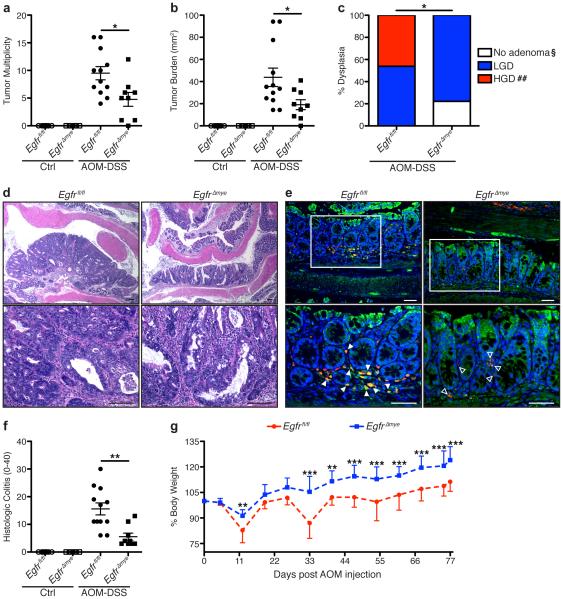
Figure 2. EgfrΔmye mice are significantly protected from tumorigenesis and dysplasia in the AOM-DSS model of colon tumorigenesis.
(a) Tumor multiplicity was assessed by gross visual inspection, utilizing a dissecting microscope. (b) Tumor burden was determined by the addition of the calculated area of each identified tumor, as assessed with an electronic caliper for both length and width. (c) Percentage of cases with either no adenoma, low-grade dysplasia (LGD), and high-grade dysplasia (HGD) determined by a gastrointestinal pathologist (M.K.W.) in a blinded manner. By Chi Square test, *P < 0.05. §P < 0.05 versus Egfrfl/fl; ##P < 0.01 versus Egfrfl/fl. n = 9-12 AOM-DSS-treated animals per genotype. (d) Representative H&E-stained images from AOM-DSS-treated mice. Scale bars = 100 μm. (e) Representative immunofluorescence images of tEGFR from AOM-DSS-treated mice. Red = CD68. Green = tEGFR. Yellow = Merge. Blue = DAPI. Scale bars = 50 μm. Solid arrows indicate CD68+tEGFR+ macrophages. Open arrows indicate CD68+tEGFR− macrophages. White box indicates zoomed area. n = ≥ 3 mice per genotype assessed. (f) Histologic colitis was determined by M.K.W. (g) Percentage of initial body weight was assessed at the indicated time points. *P < 0.05, **P < 0.01, ***P < 0.001 versus Egfrfl/fl AOM-DSS by two-way ANOVA with Bonferroni post-test (ANOVA significance = P < 0.001). In (a), (b), and (f), *P < 0.05, **P < 0.01 by one-way ANOVA with Kruskal-Wallis test, followed by Mann-Whitney U test. In (a), (b), (f), and (g), n = 7-9 control and 9-12 AOM-DSS-treated mice per genotype.