Figure 5.
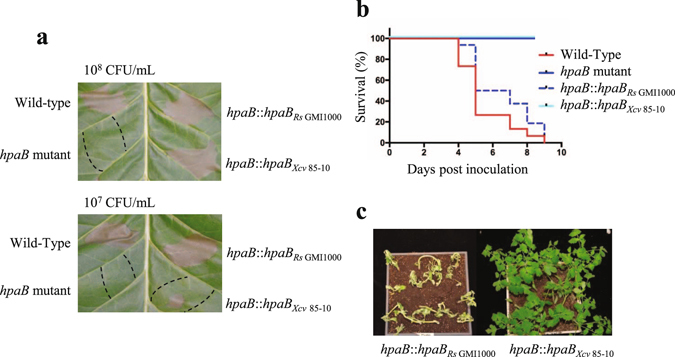
The hpaB gene from X. campestris pv. vesicatoria (Xcv) 85–10 can complement the R. solanacearum hpaB mutant to trigger an hypersensitive response (HR) on tobacco plants, but not for disease establishment on tomato plants. (a) N. tabacum leaves were infiltrated with R. solanacearum bacterial suspensions at 108 CFU/ml (upper part) and 107 CFU/ml (lower part), using the GMI1000 wild-type strain, the hpaB mutant (GMI1000 background), the hpaB mutant complemented with the hpaB gene from R. solanacearum GMI1000 and the hpaB mutant complemented with the hpaB gene from Xcv 85-10. Pictures were taken 24 h post infiltration. The dotted lines represent the infiltrated areas. Experiments were repeated three times with similar results. (b) Kaplan-Meier survival analysis of 16 tomato plants per R. solanacearum strain root inoculated. Gehan-Breslow-Wilcoxon test indicates that the wild-type strain curve (red) is significantly different from the hpaB mutant curve (blue) (p-value < 0.001), and to the curve showing the hpaB mutant complemented with the Xcv 85-10 hpaB gene (p-value < 0.001). The wild-type strain curve (red) is not significantly different from the curve showing the hpaB mutant complemented with R. solanacearum GMI1000 hpaB gene (dotted cyan) (p-value = 0.0678). Three biological repeats were performed with similar results. (c) Representative pictures showing the tomato responses six days after inoculation of the R. solanacearum hpaB mutant complemented with the hpaB genes isolated from R. solanacearum GMI1000 (left) or Xcv 85-10 (right). Experiments were repeated three times with similar results.
