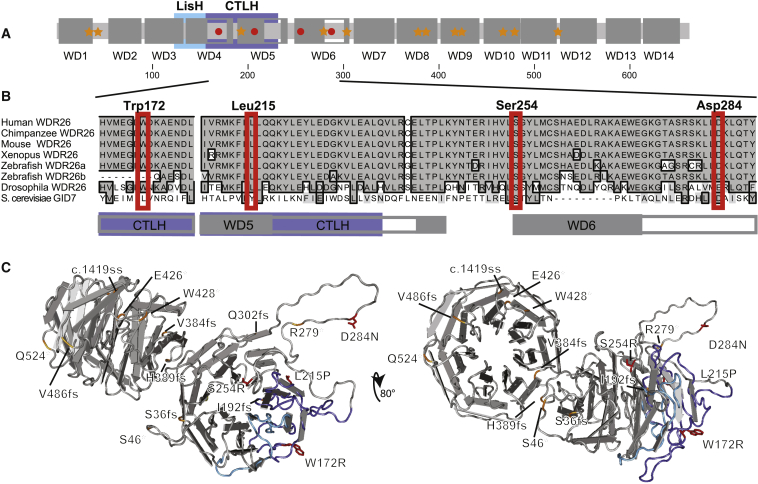
Figure 3.
Localization of WDR26 Variants
(A) Schematic domains of WDR26. Illustrated are the key domains of WDR26, which include the 14 deduced WD repeats (larger dark-gray boxes labeled WD1–WD14; insertions in the domains are noted by unfilled gray bordered segments), the LisH and CTLH homology domains (in aqua and purple, respectively), and the location of loss-of-function (orange stars) and missense (red circles) variants. Scale numbers demonstrating amino acid residues are indicated beneath.
(B) Localization of WDR26 missense variants to highly conserved residues in WD repeats near the CTLH domain. ClustalW homology alignments for human WDR26a (UniProtKB: Q9H7D7; GenBank: NP_079436.4), Chimpanzee (Pan troglodytes) WDR26 (UniProtKB: K7CSM5), mouse (Mus musculus) WDR26 (UniProtKB/Swiss-Prot: Q8C6G8.3), frog (Xenopus laevis) WDR26 (UniProtKB: A0A0H5BJW1), zebrafish (Danio rerio) WDR26a (GenBank: NP_001189371.1) and predicted WDR26b (GenBank: XP_001921656.4), fruit fly (Drosophila melanogaster) WDR26 (UniProtKB/Swiss-Prot: Q7K0L4.1), and yeast (Saccharomyces cerevisiae S288c) GID7 (UniProtKB/Swiss-Prot: P25569.2) are shown. Identical residues are in dark gray with a black outlined border. Similar residues are indicated with light-gray shading and no border. Mutated residues are noted above, and red boxes denote the position in all species. Gray and purple boxes beneath denote the WD and CTLH domain boundaries, respectively.
(C) Structural model of WDR26 with variants. Illustrated are structural models of WDR26 with a direct view of the N-terminal β propeller domain (left) and an ∼80° rotation toward the viewer to directly show the C-terminal β propeller domain (right). β sheets are illustrated as flat directional arrows. Note the organization of four β sheets into WD modules. The LisH and CTLH domains are noted by aqua and purple shading, respectively, of the protein backbone. Locations of variants are labeled and indicated; missense mutations are indicated by red shading of the residue and sidechain, and the relative position of loss-of-function alleles is noted by orange shading of the protein backbone.