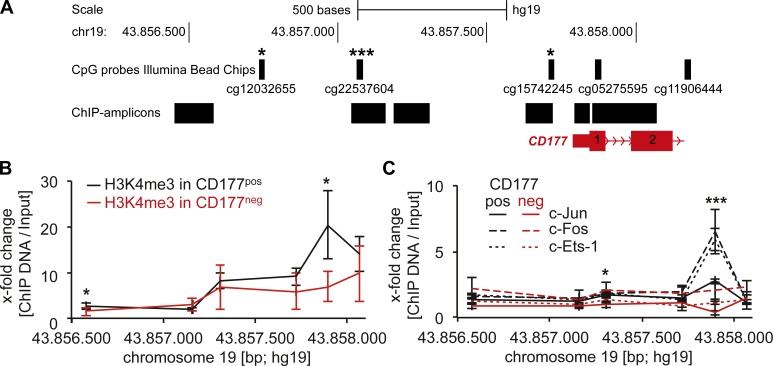
Figure 4.
Differential CpG and histone methylation, and AP-1 TF binding in the CD177 promoter of CD177neg and CD177pos neutrophil subsets. (A) Whole-genome DNA methylation analysis in CD177neg versus CD177pos neutrophils separated from CD177 bimodal individuals by magnetic cell sorting was performed using Illumina methylation chips. The localization of the 5 CpG probes (cg, CpG cluster number) within the CD177 promoter and in the CD177 gene that were present on the chip are indicated. Three of the five CpGs were significantly more methylated in the CD177neg neutrophils compared with the CD177pos subset (n = 6 different neutrophil donors). (B) ChIP assays were performed with the histone H3K4me3 mark for open chromatin in the CD177 promoter region of CD177pos (black lines) and CD177neg (red lines) neutrophils separated from a CD177 bimodal donor (n = 3 different neutrophil donors). (C) ChIP analysis for c-Jun, c-Fos, and c-Ets-1 binding was performed in CD177pos (black lines) and CD177neg (red lines) neutrophils separated from CD177 bimodal donors using specific antibodies to c-Jun, c-Fos, and c-Ets-1, respectively (n = 4 different neutrophil donors). Data in A, B, and C were analyzed using Student’s t test. *, P < 0.05; ***, P < 0.001; ±SEM is shown.