Figure 4.
Analysis of the Stability of Different mRNA Isoforms.
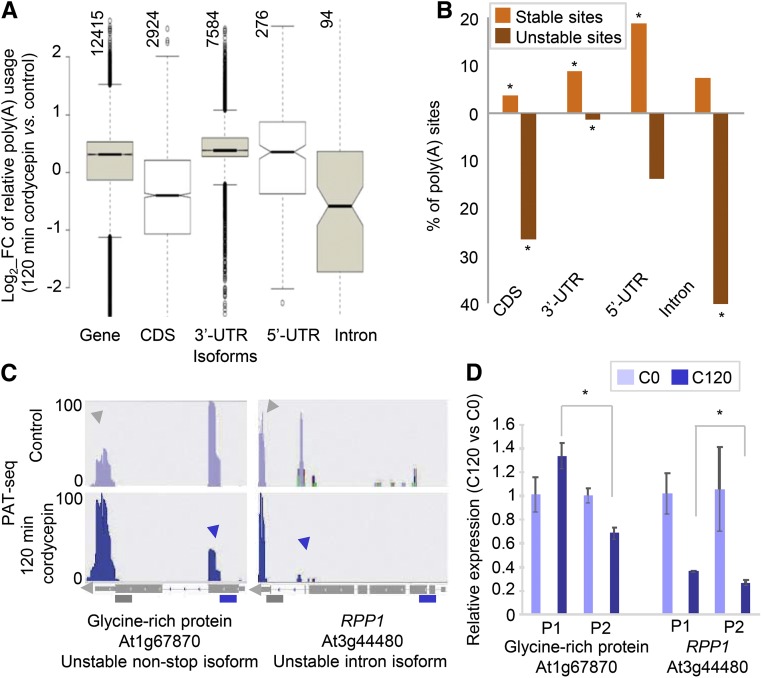
(A) Box plots showing changes in usage of different classes of poly(A) sites after 120 min of treatment with cordycepin. The relative contributions that each PAC makes to total poly(A) usage was determined as in Figure 2B. Vertically oriented numbers indicate the total number of PACs in each class.
(B) Percentage of unstable (down, <0.5×) and stable (up, >2×) mRNA isoforms. Asterisks are indicative of statistically significant differences for each isoform in each group evaluated (less or more stable), determined using the hypergeometric probability (P value < 0.0001).
(C) PAT read coverage on selected genes that illustrate differential isoform stabilities. At1g67870 provides an example of an unstable non-stop isoform and RPP1 (RECOGNITION OF PERONOSPORA PARASITICA1; At3g44480) provides an example of an unstable intronic isoform. The tracks for each gene in control and after 120 min of cordycepin treatment are shown in the upper and lower panels, respectively. The purple and gray arrows indicate the region of interest in each gene and the 3′-UTR, respectively. The read value of the scale used for each gene is indicated on the y axis (log2-transformed value).
(D) Quantitative RT-PCR analysis of poly(A) site abundance in 3′-UTR (P1) and protein-coding (P2) regions in plants grown in control and after 120 min of cordycepin conditions. Error bars represent sd between three biological replicates (pool of ∼30 whole plants per condition), and asterisks are indicative of statistically significant differences using t test (P < 0.01, three biological replicates). Purple and gray bars show the primers (as in Figure 3). C0, control conditions; C120, mRNA after 120 min of cordycepin treatment. PACs defined by fewer than 10 PATs were excluded from the analysis.