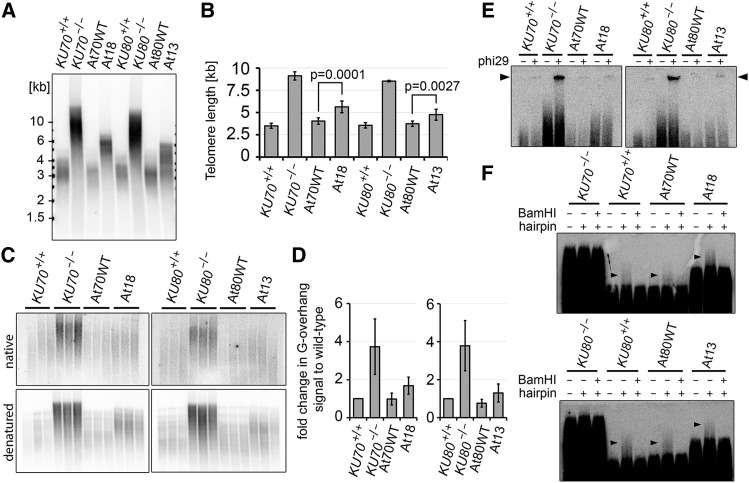
Figure 4.
Telomere Maintenance in At13 and At18 Mutants.
(A) Terminal restriction fragment analysis with Tru1I-digested genomic DNA from At18 and At13 plants and corresponding controls.
(B) Quantification of telomere length from TRF blots by TeloTool software (Göhring et al., 2014). Error bars represent sd from at least three independent samples.
(C) G-overhang analysis by the in gel hybridization technique. Restriction fragments generated by HindIII were separated by electrophoresis, and gels were first hybridized under nondenaturing conditions (top panels) than denatured and hybridized again (bottom panels).
(D) Quantification of G-overhang signal from native gels. Signals were normalized to the wild type; error bars represent sd from three independent samples.
(E) The presence of t-circles in At18 and At13 lines detected by t-circle amplification assay. Reactions without phi29 polymerase were run in parallel as a control. The signal from t-circles is indicated by the arrowhead.
(F) Integrity of blunt-ended telomeres in At18 and At13 lines determined by the hairpin ligation assay. TRFs from blunt-ended telomeres that were ligated with a hairpin migrate slower than bulk telomeres (arrowheads). The signal from blunt-ended telomeres is sensitive to cleavage of the hairpin with BamHI.