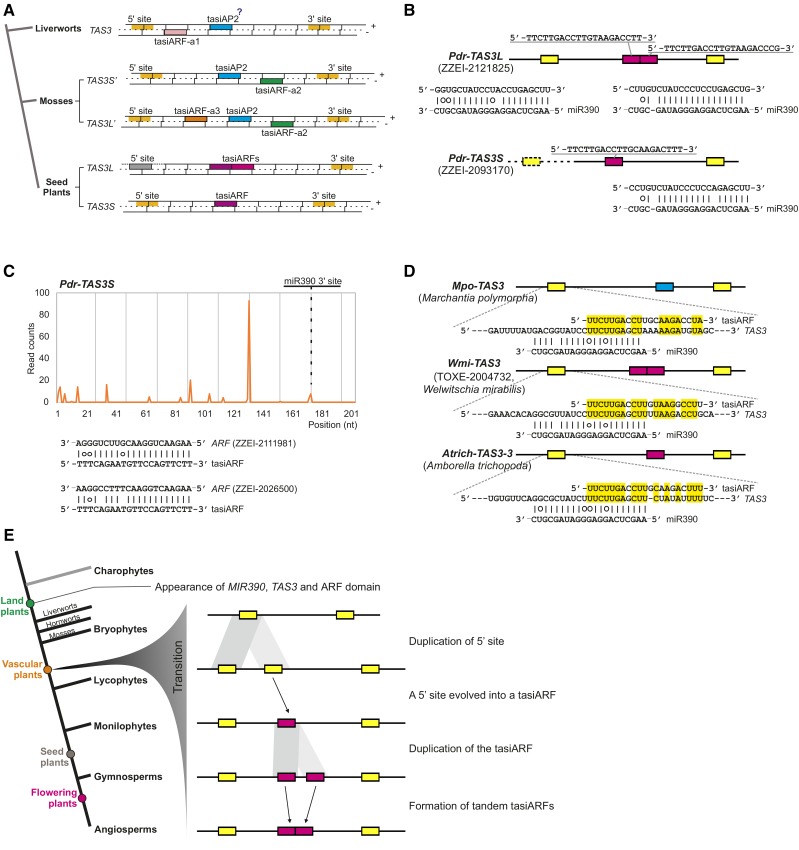
Figure 4.
The Inferred Evolutionary Progression of TAS3 Genes in Land Plants.
(A) A summary of TAS3 gene structures observed in land plants. Colored bars denote different features, as indicated; the gray 5′ miR390 site is not cleaved. The question mark indicates that the function of tasiAP2 (targeting AP2 genes) could not be validated in liverworts.
(B) Two TAS3 gene structures found in the lycophyte species P. drummondii.
(C) TAS3 transcripts produce tasiARFs to regulate ARF genes in P. drummondii. The dotted line and box denote that the identified cDNA sequence of Pdr-TAS3S was too short to include the 5′ miR390 target site.
(D) TasiARF shows sequence similarity to the region partially covering the 5′ miR390 target site of cognate TAS3 genes. Three representative TAS3 genes from different species are displayed here. Identical nucleotides between tasiARF and the region partially covering the 5′ miR390 target site are highlighted in yellow.
(E) An evolutionary model for the divergence of TAS3 genes in land plants. The tasiARF sequence originated from the duplication of the 5′ miR390 target site and the TAS3S genes (with a single tasiARF) might be the ancestor of the TAS3L genes (with two tandem tasiARFs).