Abstract
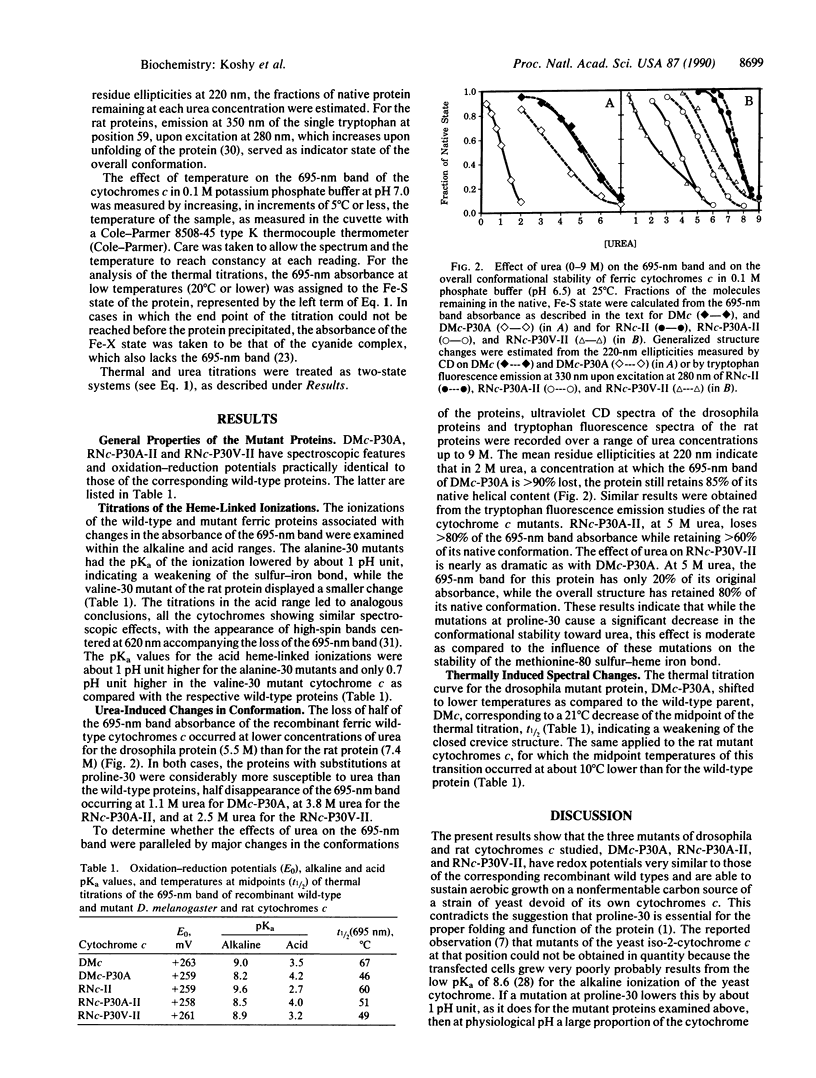
Drosophila melanogaster and rat cytochromes c in which proline-30 was converted to alanine or valine were expressed in a strain of baker's yeast, Saccharomyces cerevisiae, where they sustained aerobic growth. The mutations had no significant effect on the spectra or redox potentials but altered drastically the stability of the bond between the methionine-80 sulfur and the heme iron, as judged by four criteria: (i) the alkaline pKa values of the 695-nm band of the ferric form of the mutant proteins decreased by almost 1 pH unit as compared to the wild types; (ii) the acid pKa values increased by 0.5 to 1.2 pH units; (iii) the 695-nm band half-disappeared at temperatures 10-20 degrees C lower in the mutant proteins than in the wild types; and (iv) the 695-nm band of the mutant proteins was susceptible to concentrations of urea that had little influence on their overall structure. The valine-substituted rat cytochrome c had properties intermediate between those of the wild type and the alanine mutant. The destabilized coordinative bond is located in space a long distance from the mutation site. It is suggested that the mutations weaken the hydrogen bond between the carbonyl of residue 30 and the imino group of the imidazole of histidine-18, modifying the bonding of the heme iron by that imidazole, which, in turn, through a trans effect, weakens the bond between the heme iron and the other axial ligand, the sulfur of methionine-80. Alternatively, the effect of the mutations may be propagated allosterically along the peptide chain.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bosshard H. R. Alkaline isomerization of ferricytochrome c: lysine is not replacing methionine at the sixth co-ordination site of the haem iron. J Mol Biol. 1981 Dec 25;153(4):1125–1149. doi: 10.1016/0022-2836(81)90471-x. [DOI] [PubMed] [Google Scholar]
- Brautigan D. L., Feinberg B. A., Hoffman B. M., Margoliash E., Preisach J., Blumberg W. E. Multiple low spin forms of the cytochrome c ferrihemochrome. EPR spectra of various eukaryotic and prokaryotic cytochromes c. J Biol Chem. 1977 Jan 25;252(2):574–582. [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Carlson S. S., Mross G. A., Wilson A. C., Mead R. T., Wolin L. D., Bowers S. F., Foley N. T., Muijsers A. O., Margoliash E. Primary structure of mouse, rat, and guinea pig cytochrome c. Biochemistry. 1977 Apr 5;16(7):1437–1442. doi: 10.1021/bi00626a031. [DOI] [PubMed] [Google Scholar]
- Davis L. A., Schejter A., Hess G. P. Alkaline isomerization of oxidized cytochrome c. Equilibrium and kinetic measurements. J Biol Chem. 1974 Apr 25;249(8):2624–2632. [PubMed] [Google Scholar]
- DeLange R. J., Glazer A. N., Smith E. L. Presence and location of an unusual amino acid, epsilon-N-trimethyllysine, in cytochrome c of wheat germ and Neurospora. J Biol Chem. 1969 Mar 10;244(5):1385–1388. [PubMed] [Google Scholar]
- Dickerson R. E., Takano T., Eisenberg D., Kallai O. B., Samson L., Cooper A., Margoliash E. Ferricytochrome c. I. General features of the horse and bonito proteins at 2.8 A resolution. J Biol Chem. 1971 Mar 10;246(5):1511–1535. [PubMed] [Google Scholar]
- Dyson H. J., Beattie J. K. Spin state and unfolding equilibria of ferricytochrome c in acidic solutions. J Biol Chem. 1982 Mar 10;257(5):2267–2273. [PubMed] [Google Scholar]
- Fergusson J. E., Robinson W. T., Rodley G. A. Significance of the orientation of coordinated imidazole and benzimidazole groups in haemoprotein and vitamin B 12 structures. Aust J Biol Sci. 1972 Dec;25(6):1365–1371. doi: 10.1071/bi9721365. [DOI] [PubMed] [Google Scholar]
- George P., Lyster R. L. CREVICE STRUCTURES IN HEMOPROTEIN REACTIONS. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1013–1029. doi: 10.1073/pnas.44.10.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooley P. R., MacKenzie N. E. Pro----Ala-35 Rhodobacter capsulatus cytochrome c2 shows dynamic not structural differences. A 1H and 15N NMR study. FEBS Lett. 1990 Jan 29;260(2):225–228. doi: 10.1016/0014-5793(90)80109-v. [DOI] [PubMed] [Google Scholar]
- Hampsey D. M., Das G., Sherman F. Amino acid replacements in yeast iso-1-cytochrome c. Comparison with the phylogenetic series and the tertiary structure of related cytochromes c. J Biol Chem. 1986 Mar 5;261(7):3259–3271. [PubMed] [Google Scholar]
- Inoue S., Inoue H., Hiroyoshi T., Matsubara H., Yamanaka T. Developmental variation and amino acid sequences of cytochromes c of the fruit fly Drosophila melanogaster and the flesh fly Boettcherisca peregrina. J Biochem. 1986 Oct;100(4):955–965. doi: 10.1093/oxfordjournals.jbchem.a121808. [DOI] [PubMed] [Google Scholar]
- Kaminsky L. S., Miller V. J., Davison A. J. Thermodynamic studies of the opening of the heme crevice of ferricytochrome c. Biochemistry. 1973 Jun 5;12(12):2215–2221. doi: 10.1021/bi00736a006. [DOI] [PubMed] [Google Scholar]
- Kellis J. T., Jr, Nyberg K., Sali D., Fersht A. R. Contribution of hydrophobic interactions to protein stability. Nature. 1988 Jun 23;333(6175):784–786. doi: 10.1038/333784a0. [DOI] [PubMed] [Google Scholar]
- Limbach K. J., Wu R. Characterization of two Drosophila melanogaster cytochrome c genes and their transcripts. Nucleic Acids Res. 1985 Jan 25;13(2):631–644. doi: 10.1093/nar/13.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luntz T. L., Schejter A., Garber E. A., Margoliash E. Structural significance of an internal water molecule studied by site-directed mutagenesis of tyrosine-67 in rat cytochrome c. Proc Natl Acad Sci U S A. 1989 May;86(10):3524–3528. doi: 10.1073/pnas.86.10.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalit R., Schejter A. Cytochrome c: a thermodynamic study of the relationships among oxidation state, ion-binding and structural parameters. 1. The effects of temperature, pH and electrostatic media on the standard redox potential of cytochrome c. Eur J Biochem. 1973 Feb 1;32(3):492–499. doi: 10.1111/j.1432-1033.1973.tb02633.x. [DOI] [PubMed] [Google Scholar]
- Morishima I., Ogawa S., Yonezawa T., Iizuka T. Nuclear magnetic resonance studies in hemoproteins. IX. pH dependent features of horse heart ferric cytochrome c. Biochim Biophys Acta. 1977 Dec 20;495(2):287–298. doi: 10.1016/0005-2795(77)90385-3. [DOI] [PubMed] [Google Scholar]
- Northrup S. H., Pear M. R., McCammon J. A., Karplus M. Molecular dynamics of ferrocytochrome c. Nature. 1980 Jul 17;286(5770):304–305. doi: 10.1038/286304a0. [DOI] [PubMed] [Google Scholar]
- Northrup S. H., Pear M. R., McCammon J. A., Karplus M., Takano T. Internal mobility of ferrocytochrome c. Nature. 1980 Oct 16;287(5783):659–660. doi: 10.1038/287659a0. [DOI] [PubMed] [Google Scholar]
- Perutz M. F. Stereochemistry of cooperative effects in haemoglobin. Nature. 1970 Nov 21;228(5273):726–739. doi: 10.1038/228726a0. [DOI] [PubMed] [Google Scholar]
- Pettigrew G. W., Bartsch R. G., Meyer T. E., Kamen M. D. Redox potentials of the photosynthetic bacterial cytochromes c2 and the structural bases for variability. Biochim Biophys Acta. 1978 Sep 7;503(3):509–523. doi: 10.1016/0005-2728(78)90150-0. [DOI] [PubMed] [Google Scholar]
- Poerio E., Parr G. R., Taniuchi H. A study of roles of evolutionarily invariant proline 30 and glycine 34 of cytochrome c. J Biol Chem. 1986 Aug 25;261(24):10976–10989. [PubMed] [Google Scholar]
- SCHEJTER A., GEORGE P. THE 695-MMM. BAND OF FERRICYTOCHROME C AND ITS RELATIONSHIP TO PROTEIN CONFORMATION. Biochemistry. 1964 Aug;3:1045–1049. doi: 10.1021/bi00896a006. [DOI] [PubMed] [Google Scholar]
- Scarpulla R. C. Processed pseudogenes for rat cytochrome c are preferentially derived from one of three alternate mRNAs. Mol Cell Biol. 1984 Nov;4(11):2279–2288. doi: 10.1128/mcb.4.11.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schejter A., Plotkin B. The binding characteristics of the cytochrome c iron. Biochem J. 1988 Oct 1;255(1):353–356. [PMC free article] [PubMed] [Google Scholar]
- Shechter E., Saludjian P. Conformation of ferricytochrome c. IV. Relationship between optical absorption and protein conformation. Biopolymers. 1967;5(8):788–790. doi: 10.1002/bip.1967.360050812. [DOI] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Parker J. H., Inhaber E., Shipman N. A., Putterman G. J., Gardisky R. L., Margoliash E. The mutational alteration of the primary structure of yeast iso-1-cytochrome c. J Biol Chem. 1968 Oct 25;243(20):5446–5456. [PubMed] [Google Scholar]
- Smith G. M. Proton nuclear magnetic resonance studies of Rhodospirillum rubrum cytochrome. Biochemistry. 1979 Apr 17;18(8):1628–1634. doi: 10.1021/bi00575a039. [DOI] [PubMed] [Google Scholar]
- Smith J. A., Pease L. G. Reverse turns in peptides and proteins. CRC Crit Rev Biochem. 1980;8(4):315–399. doi: 10.3109/10409238009105470. [DOI] [PubMed] [Google Scholar]
- Swanson M. S., Zieminn S. M., Miller D. D., Garber E. A., Margoliash E. Developmental expression of nuclear genes that encode mitochondrial proteins: insect cytochromes c. Proc Natl Acad Sci U S A. 1985 Apr;82(7):1964–1968. doi: 10.1073/pnas.82.7.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T., Dickerson R. E. Conformation change of cytochrome c. I. Ferrocytochrome c structure refined at 1.5 A resolution. J Mol Biol. 1981 Nov 25;153(1):79–94. doi: 10.1016/0022-2836(81)90528-3. [DOI] [PubMed] [Google Scholar]
- Takano T., Dickerson R. E. Conformation change of cytochrome c. II. Ferricytochrome c refinement at 1.8 A and comparison with the ferrocytochrome structure. J Mol Biol. 1981 Nov 25;153(1):95–115. doi: 10.1016/0022-2836(81)90529-5. [DOI] [PubMed] [Google Scholar]
- Tsong T. Y. An acid induced conformational transition of denatured cytochrome c in urea and guanidine hydrochloride solutions. Biochemistry. 1975 Apr 8;14(7):1542–1547. doi: 10.1021/bi00678a031. [DOI] [PubMed] [Google Scholar]
- Wood L. C., White T. B., Ramdas L., Nall B. T. Replacement of a conserved proline eliminates the absorbance-detected slow folding phase of iso-2-cytochrome c. Biochemistry. 1988 Nov 15;27(23):8562–8568. doi: 10.1021/bi00423a009. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. DNA. 1984 Dec;3(6):479–488. doi: 10.1089/dna.1.1984.3.479. [DOI] [PubMed] [Google Scholar]


