Significance
Infertility is a global problem that afflicts 15% of couples, and in 50% of cases, the attributing factor is linked to men. Among these infertile men, 18% specifically exhibit decreased motility of sperm (asthenozoospermia). Sperm motility is dependent on the formation and functioning of the flagellum, a modified cilium used for locomotion. Cilia are present in almost every cell of vertebrates and are essential for proper organ functioning. Defects in cilia formation lead to severe syndromic diseases, termed ciliopathies, affecting numerous tissues (e.g., polycystic kidney disease), wherein male infertility is often comorbid. Advances in mouse genetics implicate several genes responsible for ciliopathies observed in humans. Here, we identify a nonsyndromic flagellum protein, TCTE1, that is required for sperm motility in mice.
Keywords: male infertility, asthenozoospermia, glycolysis, mutant mouse, testis-specific gene
Abstract
Flagella and cilia are critical cellular organelles that provide a means for cells to sense and progress through their environment. The central component of flagella and cilia is the axoneme, which comprises the “9+2” microtubule arrangement, dynein arms, radial spokes, and the nexin-dynein regulatory complex (N-DRC). Failure to properly assemble components of the axoneme leads to defective flagella and in humans leads to a collection of diseases referred to as ciliopathies. Ciliopathies can manifest as severe syndromic diseases that affect lung and kidney function, central nervous system development, bone formation, visceral organ organization, and reproduction. T-Complex-Associated–Testis-Expressed 1 (TCTE1) is an evolutionarily conserved axonemal protein present from Chlamydomonas (DRC5) to mammals that localizes to the N-DRC. Here, we show that mouse TCTE1 is testis-enriched in its expression, with its mRNA appearing in early round spermatids and protein localized to the flagellum. TCTE1 is 498 aa in length with a leucine rich repeat domain at the C terminus and is present in eukaryotes containing a flagellum. Knockout of Tcte1 results in male sterility because Tcte1-null spermatozoa show aberrant motility. Although the axoneme is structurally normal in Tcte1 mutant spermatozoa, Tcte1-null sperm demonstrate a significant decrease of ATP, which is used by dynein motors to generate the bending force of the flagellum. These data provide a link to defining the molecular intricacies required for axoneme function, sperm motility, and male fertility.
Flagella are ancient, analogous cellular structures used for locomotion and as sensory organelles present in all three domains of life (bacteria, archaea, and eukaryotes). The advantages conferred by this organelle are highlighted by the flagella’s apparent independent evolution in all three domains (1–3). Of all of the different flagella present among eukaryotes, flagella attached to gametes play a critical function in uniting gametes for fertilization and the perpetuation of a species. Mammalian spermatozoa have a specialized flagellum that contains a midpiece, principal piece, and end piece with the axoneme running along the entire length (4). The flagellum equips sperm with the capability to deliver half of the male’s genetic material to the female gamete, the oocyte. In addition to flagella, eukaryotes contain another related structure called cilia. The defining feature of flagella and cilia is the axoneme, the “9+2” microtubule arraignment of two central pairs of microtubules surrounded by nine pairs of microtubule doublets (5). The microtubule motor dynein is anchored to the outer microtubules and responsible for generating the force required to produce the beating pattern of flagella and cilia (6). The force generated by dynein causes sliding of the microtubules among each other; however, the nexin complex anchors the microtubules in place. The nexin complex [or nexin-dynein regulatory complex (N-DRC)] has been observed in transmission electron micrographs for several decades as a thin filament between outer microtubule doublets, but only recently has the N-DRC’s role in regulating flagellar function been elucidated (7). By preventing the sliding of microtubules, the nexin complex turns the force generated by dynein into a bending motion of both flagella and cilia. The N-DRC is a complex of up to 11 proteins that, in addition to linking adjacent outer doublets, is also thought to regulate the motor activity in dynein (7).
Failure to assemble or regulate flagellar function in spermatozoa leads to defective motility (asthenozoospermia) and fertility defects, including male sterility (8). Mutations in multiple genes in mouse models have been identified that disrupt the formation of the flagellum or the function of the flagellum (9, 10). For example, mutations in Akap4, Tekt3, Tekt4, and Cabyr in mice lead to structural defects in the flagellum, whereas mutations in CatSper1, Pgk2, Gapdhs, and Ldhc lead to functional defects in the flagellum, some of which cause defective metabolism or glycolysis (11–18). In men, mutations in CATSPER, DNAH1, DNAH11, and TEKT2 have been shown to lead to asthenozoospermia and sterility (19–22). Here, we report that a mutation in mouse T-Complex-Associated–Testis-Expressed 1 [Tcte1; the ortholog of the Chlamydomonas N-DRC component Drc5 (23)] leads to male sterility characterized by asthenozoospermia and decreased metabolism within sperm.
Results
Tcte1 Is a Testis-Enriched Gene Required for Male Fertility.
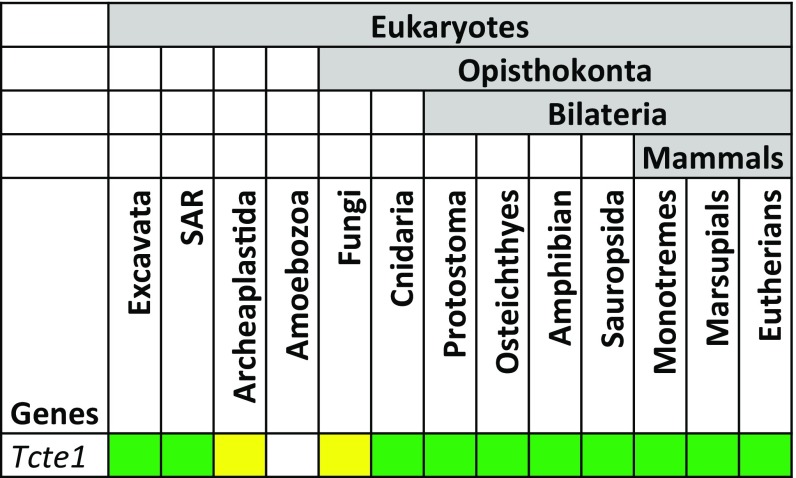
Tcte1 is an ancient, evolutionarily conserved gene present in most eukaryotic lineages with the exception of the amoebozoan lineage (Fig. S1) (24, 25). Among the basal eukaryotes, Tcte1 is present in species that contain a flagellum at some stage in the organism’s life cycle. Within the bilaterians, Tcte1 appears to be absent in Caenorhabditis elegans, an organism with nonmotile cilia and “amoeboid”-like spermatozoa (26).
Fig. S1.
Evolution of Tcte1 among eukaryotes. Tcte1 is present in most eukaryotes that contain flagella (SAR: stramenopiles, alveolates, Rhizaria). Green indicates presence of Tcte1 in most species within taxon. Yellow indicates loss in several species within taxon.
The TCTE1 protein is composed of a leucine rich repeat (LRR) similar to the LRR domain of the ribonuclease inhibitor family (Fig. 1A) with a small N-terminal portion (120 aa in the mouse) (27). The N-terminal portion is the least conserved part of the protein and varies in length among species. LRR domains have been implicated in protein–protein interactions (27).
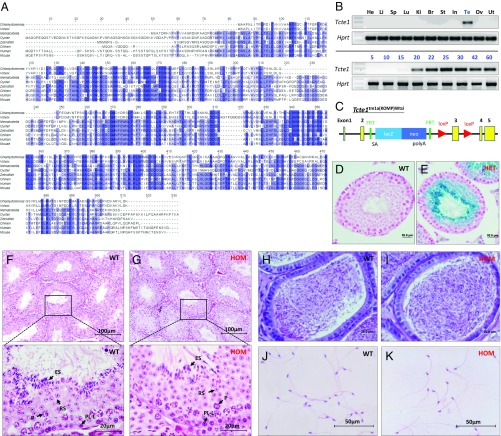
Fig. 1.
Tcte1 is a conserved testis-enriched gene required for male fertility. (A) Sequence alignment of TCTE1 proteins from several metazoans and two volvocaceaen species. Only the LRR domain is shown. (B, Top panels) RT-PCR for Tcte1 in various mouse tissues with Hprt as a control. (Bottom panels) RT-PCR from testis at various postnatal days with Hprt as a control. (C) Schematic of the Tcte1tm1a(KOMP)Wtsi (Tcte1−) allele from Wellcome Trust Sanger Institute. (D and E) β-Galactosidase staining of wild-type (D) and Tcte1+/− testis (E). (F and G) H&E-stained testis cross-sections from control (F) and Tcte1−/− mice (G). (Bottom panels in F and G: magnification of Top panels in F and G) Arrows highlight germ cells at various stages of spermatogenesis. PL-L, preleptonema-leptonema; P, pachynema; RS, round spermatid; ES, elongated spermatid. (H and I) periodic acid-Schiff (PAS)-stained cauda epididymis sections from control (H) and Tcte1−/− mice (I). (J and K) Spermatozoa from control (J) and Tcte1−/− mice (K).
We initially identified Tcte1 as a conserved testis-specific gene using a bioinformatic screen, as recently described (28). To determine the expression profile of Tcte1, we performed multitissue RT-PCR from adult mouse. RT-PCR for Tcte1 reveals a single band in testis but no other tissue (Fig. 1B, Top panels). To determine temporal expression of Tcte1, we performed RT-PCR from early postnatal day testis to capture the leading edge of the first wave of spermatogenesis. From the data, Tcte1 begins expression around postnatal day 20, which roughly corresponds to the haploid, round spermatid stage of spermatogenesis (Fig. 1B, Bottom panels). To examine the function of Tcte1 in vivo, we obtained mice containing a knockout allele of Tcte1 (Tcte1tm1a(KOMP)Wtsi, referred to as Tcte1−) from the Wellcome Trust Sanger Institute (Fig. 1C). The allele generated is part of the large mutagenesis project from the Knockout Mouse Project (KOMP) consortium (29). The KOMP Tcte1− allele contains a LacZ reporter inserted between exons 2 and 3 of endogenous Tcte1 and the third exon flanked by loxP sites (Fig. 1C). With the LacZ reporter under the control of the endogenous Tcte1 promoter, β-galactosidase staining shows expression beginning in round spermatids and persisting to elongated spermatids, which confirms the RT-PCR results (Fig. 1 D and E).
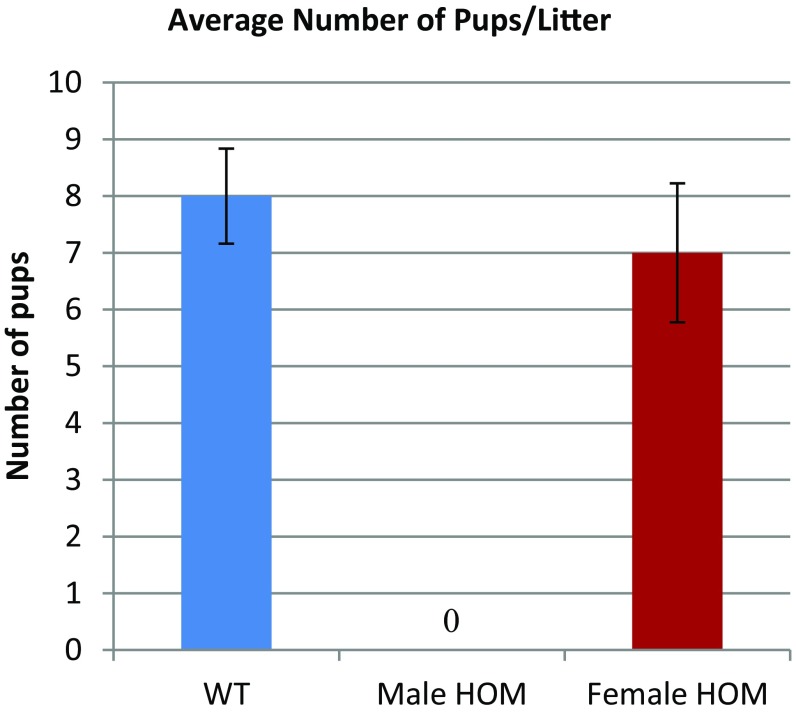
We intercrossed Tcte1+/− males and females to obtain homozygous knockout (KO) males and control littermates. Homozygous (HOM) mutant males show no overt abnormality in development or behavior. In addition, homozygous mutant females were fertile (Fig. S2, n = 5). To test male fertility, individual males (wild type and homozygous) were housed with wild-type females for 4 mo. The number of offspring per litter was recorded. Over 4 mo, five control mating pairs had, on average, 8 ± 0.84 pups per litter, whereas five KO males failed to sire any offspring despite copulation with females (i.e., copulation plugs were observed). These data demonstrate that Tcte1 is required for male fertility.
Fig. S2.
Tcte1 HOM mutant females are fertile. Bar graph showing the average number of pups/litter between wild-type males, Tcte1-null males, and Tcte1-null females (n = 5).
TCTE1 Is Required for Progressive Movement in Spermatozoa.
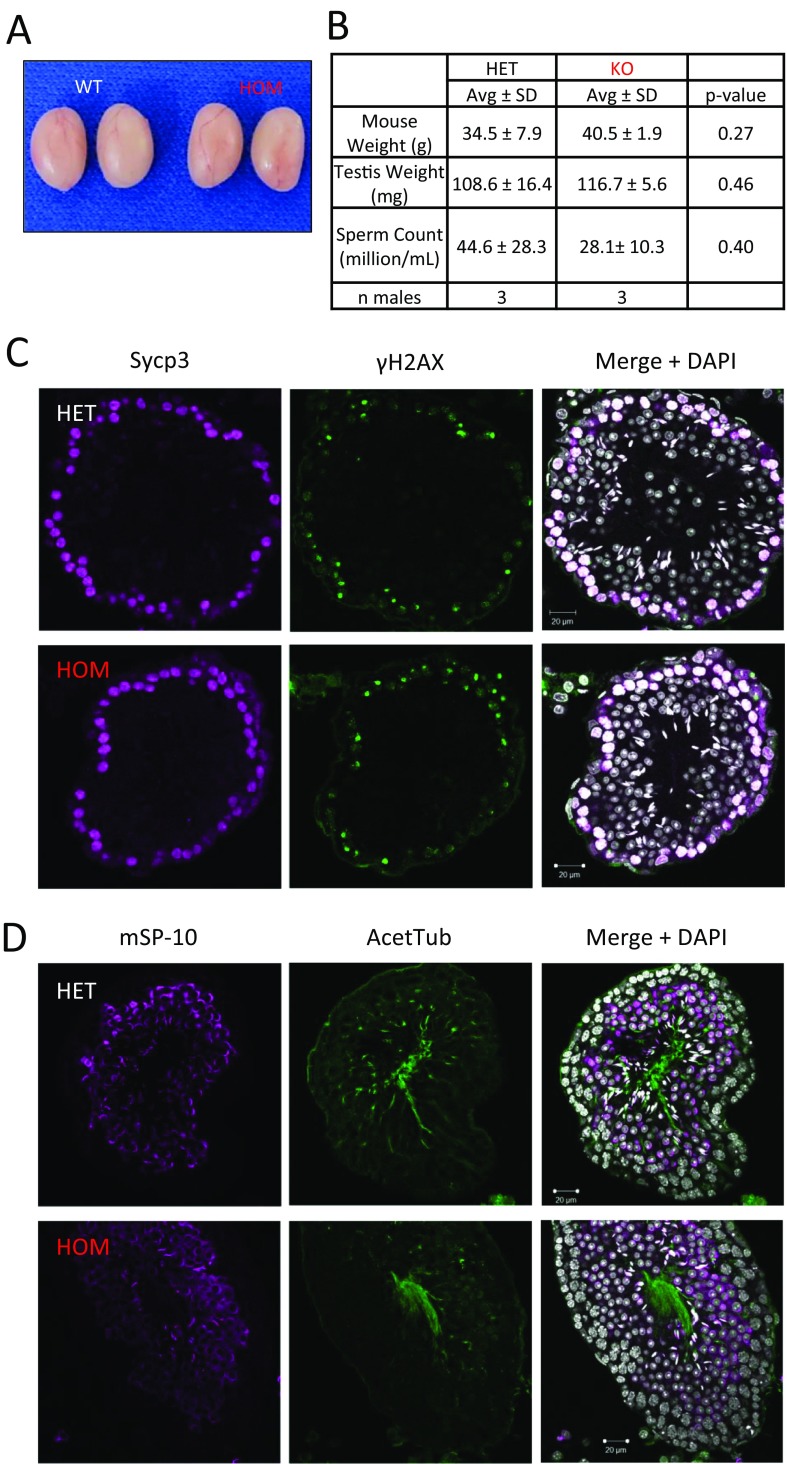
Male infertility could result from defects in spermatogenesis up to diminished fertilization ability once the spermatozoa reaches the egg (10). To determine if a block in spermatogenesis was the reason of male infertility, we examined spermatogenesis in Tcte1 null males. Gross examination of testis revealed no difference in appearance, testis weight, and sperm counts between homozygous and control littermates (Fig. S3 A and B). Using the hematoxylin–eosin (H&E) staining method, no difference was observed in spermatogenesis between wild-type control and null testes (Fig. 1 F and G). Furthermore, meiotic progression and acrosome formation did not reveal a difference between the two genotypes (Fig. S3 C and D). When examining epididymis sections, both genotypes contained tubules full of spermatozoa in the cauda and caput regions of the epididymis (Fig. 1 H and I). In addition, spermatozoa morphology appears normal in Tcte1 KOs (Fig. 1 J and K). Taken together, these observations suggest that a block in spermatogenesis is not the reason for male infertility observed in Tcte1 KOs.
Fig. S3.
Spermatogenesis appears normal in Tcte1−/− mice. (A) Testes from 9-wk-old control and KO mice. (B) Average mouse weight, testis weight, and sperm counts from three control and three KO males. (C) Meiotic markers Sypc3 and γH2AX are comparable in both control and KO testis cross-sections. (D) The acrosome caps (mSP-10) and acet-Tubulin are comparable in both control and KO testis cross-sections.
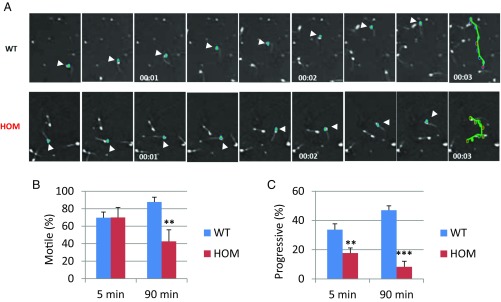
To determine the sperm quality in Tcte1−/− males, we examined sperm function using a Computer Assisted Sperm Analyzer (CASA). CASA analysis revealed decreased forwardly motile spermatozoa in KOs, a condition referred to as asthenozoospermia (Fig. 2 A–C). Despite diminished progressive movement, the sperm tails from null mice can beat; however, the range of flagellar beating is limited and appears to be asymmetric compared with that in wild type (Movies S1 and S2). Closer inspection of the sperm tails shows an inflexible midpiece (Movie S3) (30). The inflexible midpiece and asymmetric beating flagellum may account for the circular motion observed in KO sperm (Fig. 2A and Movie S3).
Fig. 2.
Tcte1 null males display asthenozoospermia. (A) Movie frames from recordings of spermatozoa from a control and homozygous mutant male. Teal: sperm head; green: path of an individual spermatozoon (arrowheads highlight sperm head). (B) Percentage of motile sperm from control and homozygous males at 5 min and 90 min (**P < 0.01). (C) Percentage of progressively motile sperm from control and homozygous males at 5 min and 90 min (**P < 0.01, ***P < 0.001).
TCTE1 Localizes to the Sperm Flagellum and Is a Component of the Dynein Regulatory Complex.
Although Tcte1 was identified in mice over 20 y ago (31), much of the functional data for Tcte1 comes from the unicellular flagellate Chlamydomonas (32). Drc5 is the Tcte1 ortholog in these unicellular green algae. Deletion and proteomic analyses in Chlamydomonas identified DRC5 as a component of the N-DRC (23, 32, 33). Nexin or “bridge” was the name for a structure observed between the outer doublets of the 9+2 arrangement from electron micrographs (32, 34). This complex interconnects the A tubule with the adjacent microtubule pair’s B to prevent sliding between tubules from the forces generated by axonemal dynein (35). Several mutants in the nexin complex disrupt the coordinated movement of the axoneme, leading to irregular flagella beating (7, 23, 32, 36, 37).
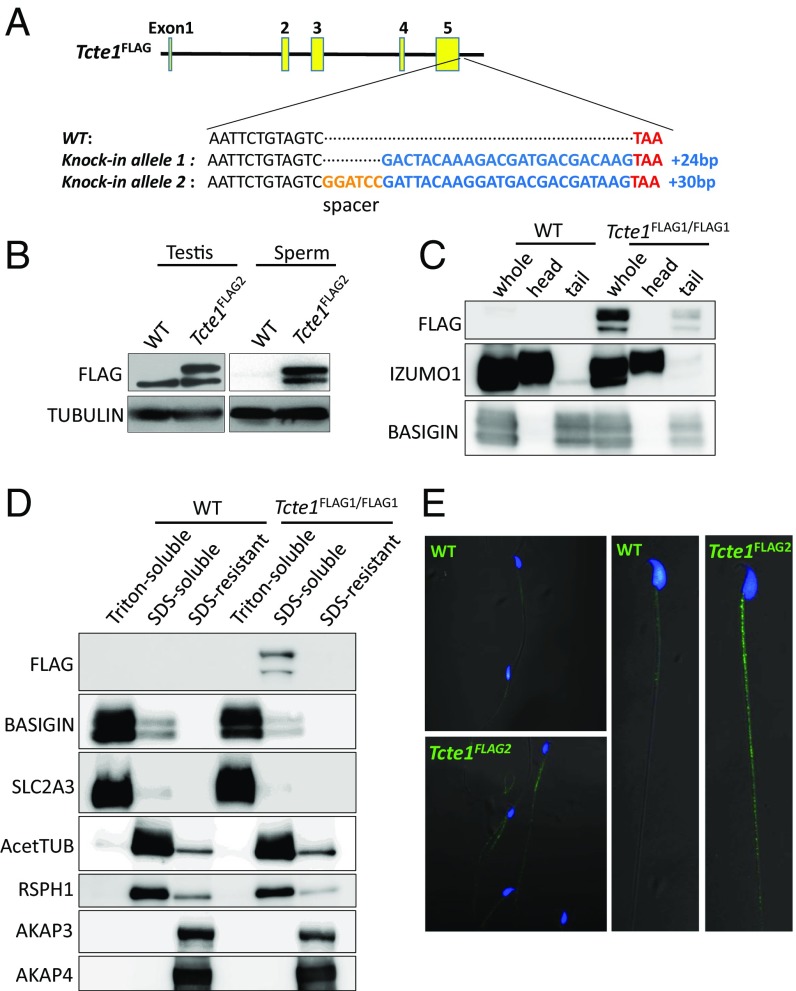
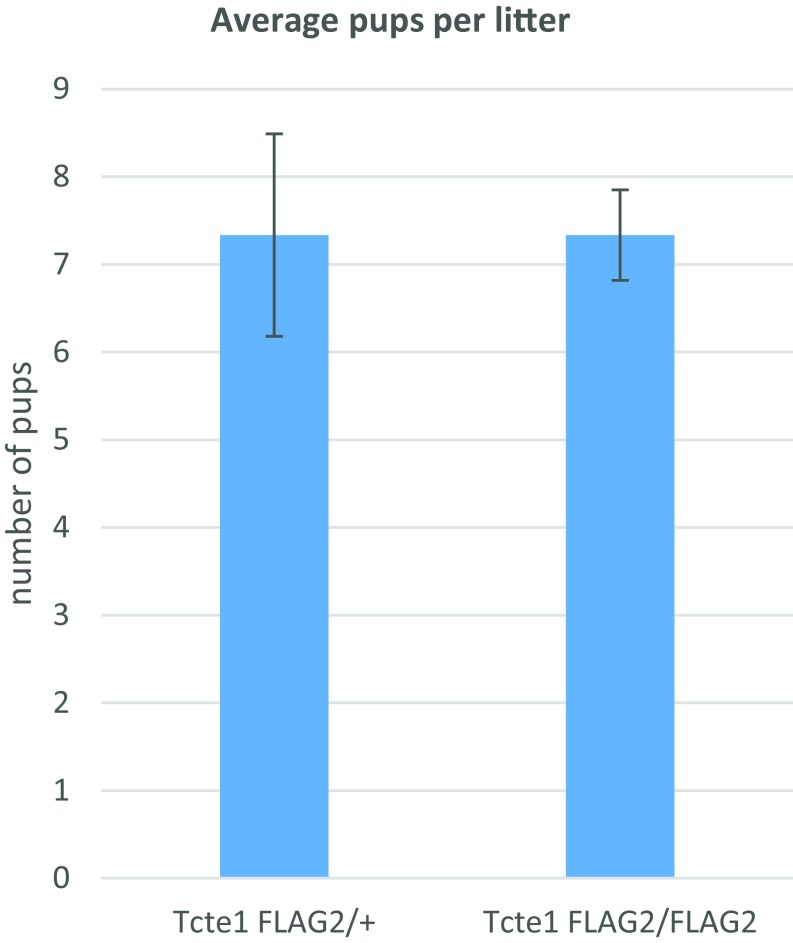
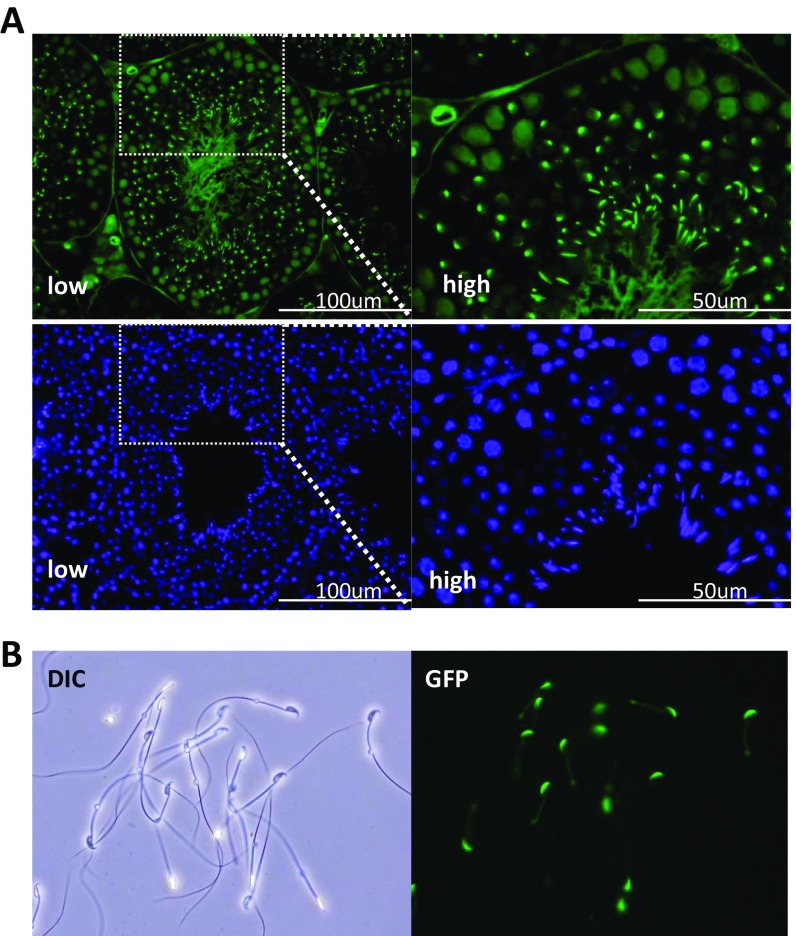
To localize TCTE1 protein, we used CRISPR/Cas9 to insert a DNA sequence into the 3′ end of the Tcte1 locus to create a TCTE1 C-terminal FLAG-tagged protein (Fig. 3A). We took an epitope tag approach as all commercially available antibodies, and two independent attempts to generate TCTE1 antibodies failed to show any promising results. To increase the chance of generating a functional TCTE1-tagged protein, we generated alleles with and without a spacer (Fig. 3A). Males homozygous for the FLAG tag (Tcte1FLAG2/FLAG2) sire pups at comparable rates to wild-type and heterozygous (Tcte1FLAG2/+) mice (Fig. S4). Also, testis cross-sections and spermatozoa from chimeric Tcte1FLAG1 generated with GFP-expressing ES cells appear normal (Fig. S5) (38, 39) (Movies S4 and S5). The motility, histology, and breeding data suggest that both Tcte1FLAG1 and Tcte1FLAG2 alleles can recapitulate TCTE1 function and indicate that the FLAG tag does not alter TCTE1 function. Western blot analysis of testis lysates using anti-FLAG antibodies detects a band at ∼50 kDa, at a position where the TCTE1-FLAG is expected to migrate, with a nonspecific band running below (Fig. 3B). When spermatozoa are fractionated into sperm heads and sperm tails, Western blot analysis using anti-FLAG antibodies also reveals a 50-kDa protein in the tail fraction (Fig. 3C). We were interested in performing immunoprecipitations (IPs) of TCTE1-FLAG to determine the TCTE1 interactome; however, our protein extracts from sperm revealed that TCTE1-FLAG is in a Triton-resistant, SDS-soluble pool (Fig. 3D). However, these results demonstrate that TCTE1 is associated with the axoneme (40). Confocal microscopy reveals the FLAG signal along the flagella of mouse spermatozoa in agreement with other work that shows that DRC5 is located along the axoneme (Fig. 3E) (32).
Fig. 3.
TCTE1 localizes to sperm tails. (A) Schematic of FLAG-tagged alleles of endogenous Tcte1 generated using CRISPR/Cas9. FLAG2 allele has a 2-aa (Gly-Ser) spacer. (B) Western blot analysis of protein extracts from testis and sperm from wild type and Tcte1FLAG2/FLAG2 mice. (C) Western blot analysis of spermatozoa fractionated into sperm heads and sperm tails from wild-type and Tcte1FLAG1/FLAG1 mice. (D) Western blot analysis of sperm fractionated into Triton X-100 soluble, SDS soluble, and SDS insoluble fractions from wild-type and Tcte1FLAG1/FLAG1 mice. (E) Immunofluorescence analysis of spermatozoa from wild-type and Tcte1FLAG2/FLAG2 mice using anti-FLAG antibody (green: anti-FLAG signal; Hoechst: blue).
Fig. S4.
Tcte1FLAG2/FLAG2 males are fertile. Breeding results from Tcte1FLAG2/+ and Tcte1FLAG2/FLAG2 males paired with wild-type females.
Fig. S5.
Tcte1FLAG1 spermatogenesis appears normal. Analysis of spermatogenesis in Tcte1FLAG1/FLAG1 males generated with ES cells expressing GFP ubiquitously and within the acrosome. (A) Cross-sections from seminiferous tubules from Tcte1FLAG1/FLAG1 chimeric males. Top panels show GFP expression ubiquitously and in the acrosome; Bottom panels show Hoechst staining. (B) Differential interference contrast and epifluorescent images of spermatozoa from Tcte1FLAG1/FLAG1 males.
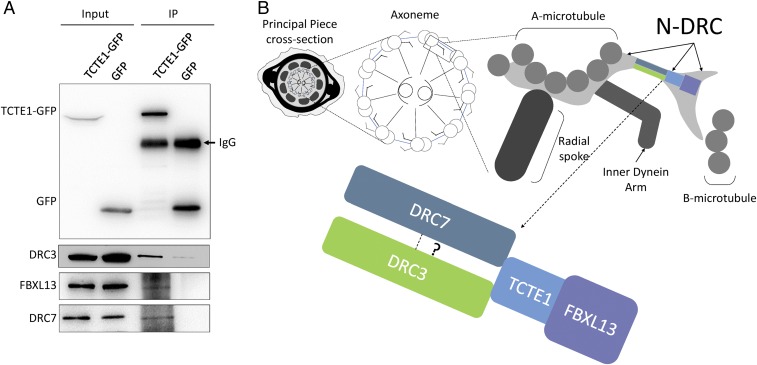
To circumvent the solubility issue of TCTE1 and the DRC from whole spermatozoa, we tested individual components of mouse DRC (DRC1-4 and 6–11) for interaction with TCTE1 (DRC5) in cell culture. GFP-tagged TCTE1, or GFP alone, was cotransfected with individual FLAG-tagged DRCs. After anti-GFP pulldowns, co-IP of DRC components was examined with anti-FLAG. Western blot analysis reveals an interaction between TCTE1 and DRC3, DRC6 (FBXL13), and DRC7 (CCDC135) (Fig. 4A) (41–43). Based on cryo-electron tomography of Chlamydomonas wild-type and mutant axonemes (23, 32), DRC5 (TCTE1), -3, -6, and -7 have been mapped proximal to each other within the DRC complex (Fig. 4B). Similar to localization of TCTE1-FLAG along the flagellum, the interaction studies are also in agreement with previous reports on TCTE1 (23, 32).
Fig. 4.
TCTE1 is a component of the dynein regulatory complex. (A) Individual DRC components were coexpressed in HEK293T cells. Immunoprecipitation of TCTE1-GFP coimmunoprecipitates DRC3, -6, and -7 (Bottom panels, third lane). (B) Model of TCTE1 localization within the DRC (adapted from ref. 23).
Sperm Axonemes Appear Normal in Tcte1-Null Sperm but Flagella Function Is Abrogated.
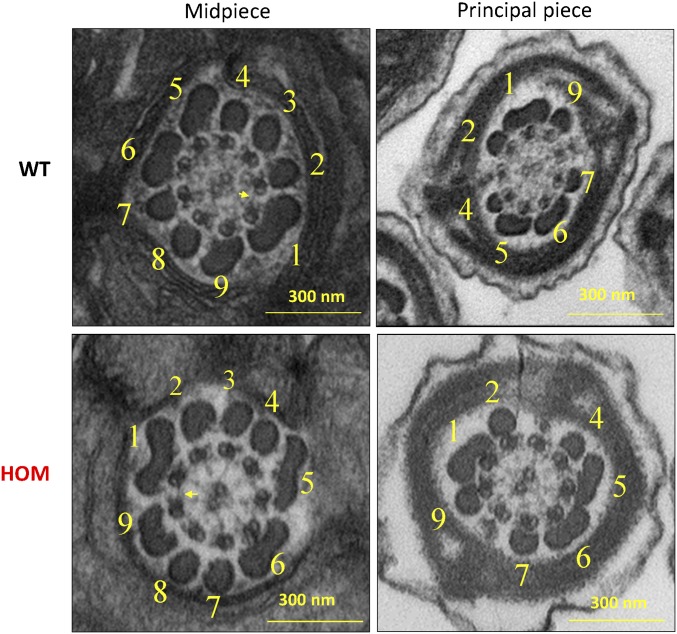
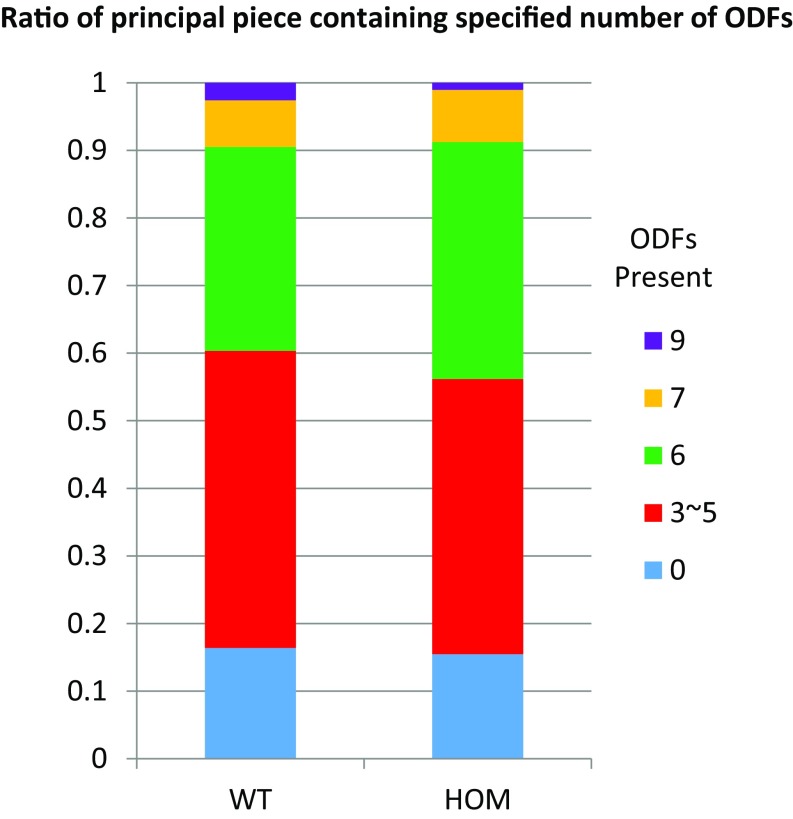
To examine whether the diminished motility of Tcte1-null sperm is due to defects in flagella organization, we examined axonemes using transmission electron microscopy (Fig. 5). Sections across the midpieces of wild-type flagella show the midpiece with mitochondria forming the outer layer followed by outer dense fibers with the axoneme at the center (4). In the principal piece, the fibrous sheath, outer dense fibers, and the axoneme are clearly distinguished. Surprisingly, the same structures are observed in electron micrographs from Tcte1−/− testis sections. The organization along the length of the Tcte1-null flagellum are comparable to wild type, and the nexin complex is visible (Fig. 5). In addition, quantification of the outer dense fibers from EM micrographs did not reveal a difference between the two genotypes (Fig. S6). This observation is consistent with work in Chlamydomonas Drc5 mutants showing little to no observable defects in axoneme organization (23, 33).
Fig. 5.
Flagellum ultrastructure appears normal. Electron micrographs showing cross-sections of the midpiece (Left panels) and principal piece (Right panels) of sperm from wild-type and Tcte1−/− mice. Numbers highlight the outer dense fibers, and yellow arrows highlight the presence of nexin links between microtubules.
Fig. S6.
The number of outer dense fibers in Tcte1−/− spermatozoa appears normal. EM cross-sections of the principal piece of sperm were quantified and categorized according to the number of outer dense fibers (ODFs) present [wild type (WT): n = 116, HOM: n = 194].
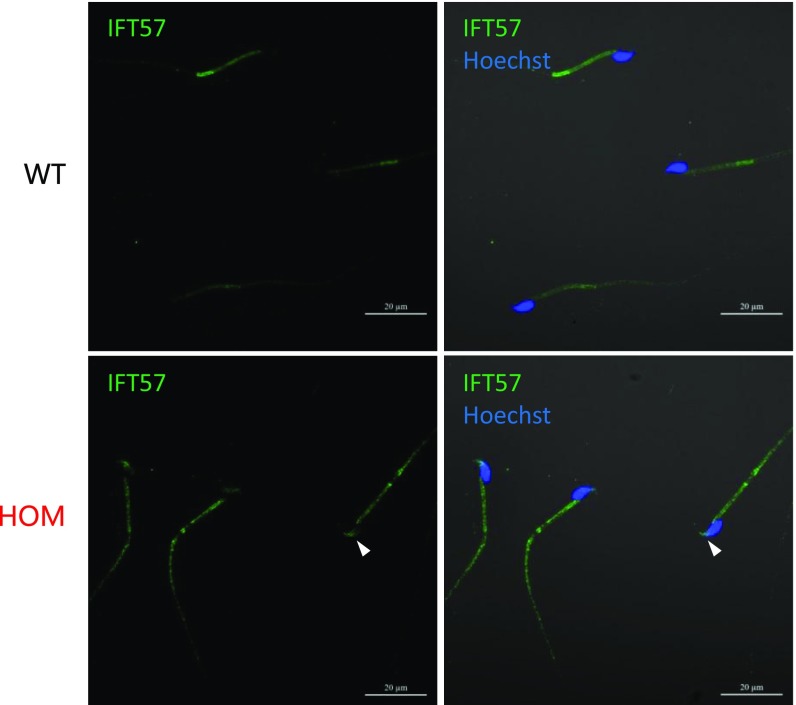
The intraflagellar transport (IFT) machinery is required for assembling the flagellum during spermiogenesis (44, 45). Immunofluorescence against IFT57 (46), an intraflagellar complex protein, shows diminished staining in wild-type spermatozoa, in agreement with previous reports (45); however, there is persistent localization of IFT57 in Tcte1-null flagellum and in sperm heads (Fig. S7, white arrowheads).
Fig. S7.
Aberrant intraflagellar transport localization. Immunofluorescence staining against IFT57 (a component of the intraflagellar transport machinery) shows persistent staining along the flagellum of Tcte1 HOM-null spermatozoa, including in the head (white arrowhead).
TCTE1 Is Required for Metabolism.
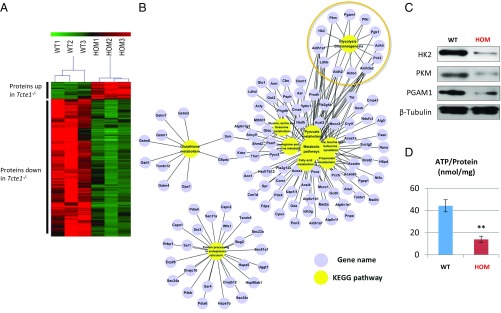
With axonemal organization appearing normal, we took a proteomics approach to whole sperm to determine if there were any differences between control and homozygous null males. Using tandem mass tag (TMT) labeling and mass spectrometry analysis, 3,697 proteins were quantified from Tcte1-null and wild-type spermatozoa. Of these proteins, 446 were differentially expressed proteins (DEPs) (Dataset S1), including 398 down-regulated proteins (DRPs, 89%) and only 49 up-regulated proteins (Fig. 6A). Components of the N-DRC were detected (Dataset S1), suggesting that TCTE1 does not regulate or stabilize N-DRC components in the mouse (Dataset S2). WebGestalt was used to identify enriched functional pathways among the DRPs (47, 48). These proteins are mostly enriched in catabolic processes (including 12 glycolytic enzymes) and protein processing in the endoplasmic reticulum (Fig. 6B). Glycolysis is a metabolic pathway that nets two molecules of ATP and pyruvate for every single molecule of glucose that is present in virtually all organisms. Interestingly, glycolysis has been shown to be crucial for producing ATP in mammalian spermatozoa (17, 18, 49–57); thus, decreased glycolytic enzyme levels could be a reason for asthenozoospermia in Tcte1-null mice. To verify results of the TMT quantification, three glycolytic enzymes—hexokinase-2 (HK2), pyruvate kinase (PKM), and phosphoglycerate mutase 1 (PGAM1)—were evaluated using commercially available antibodies for Western blot analysis. As shown in Fig. 6C, Western blot and TMT quantification showed consistent results. When we quantified ATP amount from both control and Tcte1-null spermatozoa, we discovered that ATP generation from Tcte1-null spermatozoa is significantly lower than from wild type (Fig. 6D).
Fig. 6.
Glycolysis is decreased in Tcte1-null sperm. (A) Heat map from three independent proteomic analyses of sperm from wild-type and Tcte1-null mice. Red: proteins up-regulated; green: proteins down-regulated. (B) Grouping of proteins down-regulated in Tcte1-null sperm according to metabolic pathways. (C) Measured levels of ATP between wild-type and Tcte1-null sperm (n = 3). (D) Western blot analysis of three glycolytic enzymes from wild-type and Tcte1-null sperm.
Discussion
We showed that Tcte1 is an evolutionarily conserved gene that is present in nearly all eukaryotes that contain a flagellum or motile cilium at some point in their life cycle and appears to be lost in species that lack these cellular organelles. The evolutionary gene loss is highlighted by C. elegans, an organism that contains nonmotile cilia but no homologs to TCTE1 or several other N-DRC genes (26). In the mouse, Tcte1 is a testis-enriched gene that begins expression during the haploid phase of spermatogenesis. During the haploid phase of spermatogenesis, referred to as spermiogenesis, several flagellar proteins are translated and begin to form the axoneme that will eventually be surrounded by outer dense fibers, the fibrous sheath, and mitochondria (4). Although the N-DRC is present in motile cilia and several cell types of the mouse contain motile cilia, no other observable defect is apparent in Tcte1-null mice. There are several possible reasons for this observation. It is likely that TCTE1 became specialized for the function of sperm flagella and that there is either a paralogue or an analog that replaces TCTE1 in somatic cilia.
In the absence of TCTE1, male mice are sterile due to asthenozoospermia (sperm with diminished progressive motility). CASA analysis demonstrates that the diminished motility is due to aberrant flagellar function, with sperm tails contracting toward one side and generating a circular path for sperm. The N-DRC is thought not only to prevent microtubule sliding, but also to coordinate dynein motor function (58). Perhaps in the absence of TCTE1, either there is sliding of microtubules among each other or uncoordinated dynein motor function throughout the flagellum that prevents the dynein forces from being translated to bending of the flagellum. TCTE1 contains a conserved LRR domain, which is known to mediate protein–protein interactions, at the C terminus with no other domains of catalytic function. It is possible that TCTE1 binds a regulator or a structural protein of the N-DRC, allowing for the N-DRC to maintain proper regulation of the axoneme. However, despite the absence of Tcte1, the axoneme ultrastructure appears normal, and the N-DRC complex appears to be present. This is not an unusual finding as several N-DRC mutants (including Drc5 mutants) in Chlamydomonas can form the axoneme, the nexin link between outer doublets, and even show flagellar beating (23, 33). Perhaps there is a hierarchy in assembly of the N-DRC, as various ranges of phenotypes in Chlamydomonas suggest, and DRC5 is more downstream of the pathway that is required for the finer movements and coordination of the dynein power stroke.
Localization of TCTE1-FLAG in the flagellum using immunofluorescence and in vitro interactions of TCTE1 with three N-DRC components (DRC3, -6, and -7) support TCTE1 as an evolutionarily conserved component of the dynein regulatory complex. Immunoprecipitation would be advantageous in probing a more complete proteome of TCTE1; however, immunoprecipitation has been hampered because much of TCTE1 is present in the insoluble fraction from sperm extracts. Proteomic work on sperm extracts will be beneficial in overcoming TCTE1’s insolubility and determining whether TCTE1 interacts with additional DRCs or non-DRC proteins hypothesized to reside within the nexin link (32).
The main driving force of flagellar movement is generated by the AAA ATPase of dynein (6). We showed that ATP levels are lower in Tcte1-null sperm compared with wild type, which could account for the decreased motility. Perhaps as the pool of ATP is hydrolyzed, Tcte1-null sperm lose their motility and ultimately stop when ATP levels are extinguished. Why the levels of ATP would be decreased is not clear. Of note, TCTE1 has not been shown to be a member of the glycolytic pathway or involved in the regulation of this pathway. It is possible that the effect on ATP levels is indirect. For example, perhaps dynein activity is increased, leading to increased metabolism of ATP in Tcte1-null sperm. The Tcte1-null phenotype is also reminiscent of Gapdhs-null mice, where ATP levels are down to a similar extent as Tcte1-null males and sperm motility is diminished (17). GAPDHS is the testis-specific ortholog of somatic GAPDH; in addition, there are other testis-specific glycolytic paralogues. Glycolysis is essential for motility because much of the ATP used by dynein is generated via the glycolytic pathway and not through the electron transport chain (17, 18, 49–57). It is tempting to speculate that TCTE1 influences the glycolytic pathway; however, glycolysis is thought to occur at the fibrous sheath, as many glycolytic enzymes are covalently attached to that structure (40, 57). The IFT machinery may link TCTE1 and the fibrous sheath. IFT is essential for formation of the flagellum (45, 59, 60) during spermiogenesis but is absent in spermatozoa. As there is persistent IFT57 localization in Tcte1-null spermatozoa, perhaps IFT is abrogated, leading to subtle defects in fibrous sheath assembly that we have been unable to detect. Further work is required to understand how TCTE1, glycolysis, and the axoneme function together to generate the motility required to deliver the haploid genome of male mice to the ovum. Here, we demonstrate that the sperm flagellum is an excellent cellular site to study genes required for flagellum and cilium formation and function. With more sophisticated genetic analyses using CRISPR/Cas9, we hope to untangle the molecular intricacies of the flagellum.
Materials and Methods
Animals.
Tcte1tm1a(KOMP)Wtsi/+ (referred to as Tcte1+/−) mice were obtained from the KOMP consortium. The null allele was generated using JM8A3.N1 ES cells, diphtheria toxin A (DTA) containing backbone, and homology arms 5.8 kb upstream and 3.9 kb downstream of Tcte1 (Mouse Genome Informatics: 98640). Tcte1FLAG1 and Tcte1FLAG2 were generated as described below. All mice were housed in a specific pathogen-free animal facility with a light:dark cycle of 12:12. All animals in this study were approved by the Institutional Animal Care and Use Committees of Nanjing Medical University, Nanjing, China; Baylor College of Medicine, Houston; and Osaka University, Osaka.
RT-PCR.
Mouse cDNA was prepared from multiple adult tissues of C57BL6J/129SvEv hybrid mice and testes from 5- to 60-d-old mice.
Generation of Tcte1tm2(Maik) (Referred to as Tcte1FLAG1) Mice by CRISPR/Cas9 (ES Method).
Single-guide RNA (sgRNA)- and Cas9-expressing plasmid and reference plasmid for homologous recombination of FLAG were transfected into EGR-G01 ES cells as previously described (61). Cas9- and sgRNA-expressing plasmid was constructed in pX459 to target the C-terminal of Tcte1 (target sequence: 5′-gaattctgtagtctaagatg-3′). The reference plasmid was constructed in pBluescript II SK (+) vector containing 500 bp of the homology arms 5′ and 3′ on either side of the FLAG sequence. The 1-kb wild-type allele was first amplified and then cloned into the XbaI-XhoI sites. The FLAG sequence was inserted by replacing the 3′ arm of the wild-type allele using EcoRI and XhoI. After establishment of transfected ES cell clones, genotyping was performed by PCR (Dataset S3). The mutant ES cell clones were injected into eight-cell ICR (Institute of Cancer Research) embryos, and the chimeric blastocysts were transported into the uteri of pseudopregnant females. Tcte1FLAG1 male mice were obtained after mating chimeric mice and wild-type female mice.
Generation of Tcte1tm3(Mngx) (Referred to as Tcte1FLAG2) Mice by CRISPR/Cas9.
Cas9 mRNA was produced and purified as previously described (62). In brief, Cas9 plasmid (Addgene) was linearized with AgeI and then purified using a MinElute PCR Purification Kit (Qiagen). Cas9 mRNA was produced by in vitro transcription using a mMESSAGE mMACHINE T7 Ultra Kit (Ambion) and purified using a RNeasy Mini Kit (Qiagen) per the manufacturer’s instructions. sgRNA was designed near the stop codon of Tcte1 (Dataset S3). The two cDNA oligos were annealed and then ligated into BsaI-digested pUC57-T7-sgRNA vector. sgRNA plasmid was linearized with DraI and then purified using a MinElute PCR Purification Kit (Qiagen). sgRNA was produced using the MEGAshortscript Kit (Ambion) and purified using the MEGAclear Kit (Ambion) per the manufacturer’s instructions. The donor ssDNA sequence shown in Dataset S3 includes a 56-bp left arm and a 58-bp right arm (with a stop codon) and 1× Flag sequence with six additional nucleotides (GGATCC). Mouse zygotes were coinjected with Cas9 mRNA (25 ng/µL), sgRNA (15 ng/µL), and donor ssDNA (1 μM).
Histological Analysis.
For β-galactosidase staining, testes were fixed overnight at 4 °C (100 mM phosphate buffer, pH 7.3; 2% paraformaldehyde; 0.2% glutaraldehyde) and then rinsed three times at room temperature (100 mM phosphate buffer, pH 7.3; 2 mM MgCl2; 0.01% sodium deoxycholate; 0.02 Triton x-100). Staining was done using X-gal buffer (100 mM phosphate, pH 7.3; 2 mM MgCl2; 5 mM K3Fe(CN)6; 5 mM K4Fe(CN)6; 0.01% sodium deoxycholate; 0.02% Nonidet P-40; 1 mg/mL X-gal) for 12–24 h at room temperature protected from light. Samples were rinsed three times with PBS for 10 min per wash and then stored in 10% neutral buffered formalin at 4 °C until ready to process and embed. The Baylor College of Medicine Histology Core performed processing and embedding. Paraffin blocks were sectioned at 10 μm. Slides were cleared with Histo Clear, rehydrated, and counterstained with Nuclear Fast Red. Stained slides were washed with dH2O, dehydrated, and mounted with Permount or VectaMount.
For H&E staining, testes were fixed in modified Davidson’s Fluid (30% of a 37–40% stock solution of formaldehyde, 15% ethanol, 5% glacial acetic acid, and 50% distilled H2O) and embedded in paraffin. Sections were cut at a 5-μm thickness. Sections were then deparaffinized, rehydrated, stained with H&E, dehydrated, and mounted.
For periodic acid-Schiff (PAS) staining, epididymis was fixed in Bouin’s Fixative, processed, and embedded in paraffin. Sections were cut at a 10-μm thickness. Sections were then deparaffinized, rehydrated, stained with Periodic Acid-Schiff’s Reagent, counterstained with hematoxylin, dehydrated, and mounted.
Ultrastructural examination has been described previously (63). Briefly, 4% (vol/vol) glutaraldehyde-fixed sperm were postfixed with 2% (wt/vol) OsO4 and embedded in Araldite. Ultrathin sections (80 nm) were stained with uranyl acetate and lead citrate and analyzed by EM (JEM.1010, JEOL).
Sperm Analysis.
Sperm from Tcte1−/− and from control littermates (Tcte1+/− and wild type) were extracted and incubated in human tubal fluid (HTF) media (Millipore) supplemented with 10% FBS at 37 °C. Sperm samples were diluted and analyzed using Hamilton Thorne’s Ceros II system.
Head–Tail Separation.
Spermatozoa from the cauda epididymis were suspended in 1 mL PBS and sonicated to separate tails from heads (10 s, three times, Sonifier SLPe, Branson Ultrasonics). The sample was centrifuged at 10,000 × g for 5 min. The pellet was then resuspended with 200 μL PBS, mixed with 1.8 mL of 90% Percoll solution (GE Healthcare) in PBS, and centrifuged at 15,000 × g for 15 min. The bottom and top layer of the Percoll solution contains heads and tails, respectively. The separated sample was diluted 1:5 with PBS and centrifuged at 10,000 × g for 5 min. After washing twice with PBS, the collected samples were dissolved in sample buffer for immunoblot analysis.
Fractionation of Spermatozoa.
Spermatozoa were suspended in 1% Triton X-100 lysis buffer (50 mM NaCl, 20 mM Tris⋅HCl, pH 7.5, protease inhibitor mixture) and incubated at 4 °C for 2 h. The sample was centrifuged at 15,000 × g for 10 min to separate the Triton-soluble fraction (supernatant) and the Triton-resistant fraction (pellet). The pellet was resuspended in 1% SDS lysis buffer (75 mM NaCl, 24 mM EDTA, pH 6.0) and incubated at room temperature for 1 h. The sample was centrifuged at 15,000 × g for 10 min to separate SDS-soluble fraction (supernatant) and SDS-resistant fraction (pellet). The pellet was dissolved in sample buffer. Antibodies used: rabbit anti-RSPH1 1:500 (64); rat monoclonal anti-SLC2A3 1:500 (65); rat monoclonal anti-IZUMO 1:500 (66); goat anti-BASIGIN 1:500 (Santa Cruz sc-9757); rabbit polyclonal anti-FLAG 1:500 (MBL PM020); and mouse anti-AcetTUB 1:1,000 (Sigma T7451).
Immunofluorescence.
For testis cryosections, immunostaining was performed as previously described (67). Primary antibodies used include: mouse anti-AcetTubulin 1:1,000 (Sigma); guinea pig anti-mSP10 1:200 (68); mouse anti-gH2AX 1:1,000 (Millipore); rabbit anti-SYCP3 1:1,000 (abcam). Secondary antibodies used include AlexaFluor 594 anti-rabbit, AlexaFluor 488 anti-mouse, and AlexaFluor 594 anti-guinea pig.
For spermatozoa, samples were spread onto microscope slides and allowed to air-dry. The samples were then fixed with 4% paraformaldehyde in PBS for 20 min. After three 10-min washes with PBS, heat-induced antigen retrieval was carried out by boiling the slides in 10 mM citrate buffer (pH 6.0) with a microwave oven for 10 min. After three 10-min washes with PBST (0.1% Triton X-100 in PBS), the slides were blocked with 5% BSA diluted in PBST for 1 h and then incubated with anti-FLAG mAb (1:1,000 in 5% BSA (MBL) overnight. After incubation with secondary antibody at room temperature for 2 h, the slides were incubated with Hoechst 33342 for 5 min. Finally, the slides were washed in PBS and then mounted with VectaShield or Immu-Mount. Slides were viewed with an LSM700 confocal microscope (Carl Zeiss AG).
Immunoprecipitation from Cell Culture Extracts.
HEK293T cells were transfected with individual DRC expression plasmids using Lipofectomine 2000. Two days after transfection, cells were lysed with Pierce IP Lysis Buffer (Thermo Scientific) supplemented with 1% (vol/vol) protease inhibitor mixture (Selleck) for 40 min at 4 °C and then clarified by centrifugation at 12,000 × g for 20 min. The lysates were mixed accordingly (TCTE1-GFP or GFP with individual DRCs) and precleared with 20 µL Protein A/G magnetic beads (Selleck) for 1 h at 4 °C. Precleared lysates were incubated overnight with anti-GFP antibody at 4 °C. Lysates were then incubated with 50 µL Protein A/G magnetic beads for 3 h at 4 °C. The beads were washed three times with IP Lysis Buffer and eluted with Pierce IgG Elution Buffer (Thermo Scientific). Samples were boiled in SDS loading buffer before SDS/PAGE. Tcte1-Plvx-AcGFP1-N1 and negtive control Plvx-AcGFP1-N1 were purchased from Vazyme Biotech. The DRCs were cloned into pcDNA3.0 with 3 × FLAG tag inserted (primer sequences in Dataset S3).
Proteomic Digestion and TMT Labeling.
Proteomic digestion was performed as described (69). Epididymal sperm were lysed in protein extraction buffer [8 M urea, 75 mM NaCl, 50 mM Tris, pH 8.2, 1% (vol/vol) EDTA-free protease inhibitor, 1 mM NaF, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 1 mM PMSF]. Protein concentration was measured using the Bradford assay. Cysteine residues were reduced with DTT at a 5-mM final concentration for 25 min at 56 °C followed by alkylation in 14 mM iodoacetamide for 30 min at room temperature in the dark. Unreacted iodoacetamide was quenched with DTT for 15 min. Lysates were then diluted down to 1.6 M urea with 25 mM Tris, pH 8.2. CaCl2 was added to a final concentration of 1 mM and digested overnight at 37 °C with trypsin in a 1:200 enzyme to protein ratio. For TMT labeling, purified peptides were reconstituted in 200 mM triethylammonium bicarbonate (TEAB). Twenty-two microliters of TMT reagent (Pierce) was added to 45 µg peptides (measured by A280) and allowed to react for 1 h at room temperature. Wild-type samples were labeled with 126, 128, and 130 labels and the KO with 127, 129, and 131 labels. The reaction was quenched for 15 min by addition of 5% hydroxylamine. After TMT labeling, all six samples were combined and purified using an OASIS HLB 1-cc Vac cartridge (Waters) and then lyophilized.
High-pH Reverse Phase Fractionation.
High-pH reverse phase fractionation was performed as described (70). The peptide mixture was resuspended in 90 µL buffer (10 mM ammonium acetate, pH 10) and loaded onto a XBridge BEH130C18 column (2.1 × 150 mm, 3.5 μm; Waters) with the UltiMate 3000 HPLC systems at a flow rate of 200 μL/min. A total of 120 fractions were collected with a 60-min gradient: 0–7% buffer B (90% ACN/10 mM ammonium acetate, pH 10) for 3 min, 7–42% B for 40 min, 42–70% B for 12 min, followed by 70% buffer B for 5 min, monitored at 214 nm based on the UV-light trace. These 120 fractions were then pooled to generate the final 30 fractions using a nonadjacent pooling scheme (e.g., 1, 31, 61, 91). The fractions were then dried in a vacuum.
LC-MS/MS Analysis.
For MS analyses, peptides were resuspended in 0.1% formic acid and analyzed using a LTQ OrbitrapVelos mass spectrometer (Thermo Finnigan) coupled with a Proxeon Easy-nLC 1000. Peptides were first loaded onto the trap column (75 μm × 2 cm, Acclaim PepMap100 C18 column, 3 μm, 100 Å; DIONEX) at a flow rate of 10 μL/min and then transferred to a reverse-phase microcapillary column (75 μm × 2 5 cm, Acclaim PepMap RSLC C18 column, 2 μm, 100 Å; DIONEX) at a flow rate of 300 nL/min. The HPLC solvent A was 0.1% formic acid (FA), and the solvent B was 100% acetonitrile (ACN) with 0.1% FA. The following linear gradient was used: 3–8% buffer B for 3 min, 8–29% buffer B for 176 min, 29–41% buffer B for 15 min, 41–100% buffer B for 1 min, and 100% buffer B for 10 min. Peptide analysis was performed on a LTQ OrbitrapVelosin using the data-dependent acquisition mode. An MS survey scan was obtained for the m/z range 400–1,800 at a resolution of 60,000 and a low-energy MS/MS scan of every precursor in the linear ion trap (collision-induced dissociation) followed by a higher energy MS/MS scan in the octopole collision cell [higher energy collision dissociation (HCD)] was acquired from the survey scan for the eight most intense ions (as determined by X-calibur mass spectrometer software in real time). Dynamic mass exclusion windows of 60 s were used, and siloxane (m/z 445.120025) was used as a lock mass.
Protein Identification and Quantification.
All raw files were searched with MaxQuant software (version 1.2.2.5) using the UniProt human proteome database (version 2014_08) (71). The reverse strategy was used to estimate the false discovery rate (FDR). Both the peptide and protein FDR were set to 0.01. Enzyme specificity was set to Trypsin/P, and the maximum missed cleavage sites permitted was two. The minimum peptide length required was six. Carbamidomethylation on cysteine, and TMT reagent adducts on lysine and peptide amino termini were set as fixed modifications. Methionine oxidation and acetyl (protein N-term) were set as variable modifications. The mass tolerance for precursor ions was set to 20 ppm at the first search as applied in MaxQuant for initial mass recalibration. For the main search, the mass tolerance for precursor ions was set to 6 ppm. The mass tolerance for MS/MS fragment ions was set to 0.5 Da. Relative protein abundance ratios between the two groups were calculated from TMT reagent reporter ion intensities from HCD spectra according to the Libra algorithm (72). Statistical significance was determined using the unpaired two-tailed Student’s t test. Proteins with a P value less than 0.05 and a fold change greater than 1.5 were considered as differentially expressed between groups. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE (73) partner repository with the dataset identifier PXD005343.
Selected Reaction Monitoring Assay Configuration.
Selected reaction monitoring (SRM) assays were configured based on experimental tandem MS (MS/MS) data for the peptide mixtures from TCTE1. Two unique peptides with the best flyer properties and intense fragmentation patterns, QVSALGSSSTGPTSLK and TILEFDLR, were selected and the precursor-to-fragmention transitions were tested by SRM measurements on a TSQ Quantum Ultra triple quadrupole mass spectrometer (Thermo Scientific).
Western Blot Analysis of Tcte1FLAG Sperm Extracts.
Protein extracts were prepared using lysis buffer [7 M urea, 2 M thiourea, 2% (wt/vol) DTT] containing a 1% (wt/vol) protease inhibitor mixture (Pierce Biotechnology). Western blot analysis was performed as described previously (74) with slight modifications. Briefly, the proteins were separated by SDS/PAGE and transferred onto a polyvinylidene difluoride membrane. The membranes were blocked with 5% nonfat milk in tris-buffered saline and polysorbate 20 (TBST) solution for 2 h at room temperature and incubated overnight at 4 °C with primary antibodies. The membranes were washed with TBST buffer three times and incubated at room temperature for 2 h with secondary antibodies. The signals from the detected proteins were visualized using SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific).
Measurement of Sperm ATP in Vitro.
Sperm samples were washed twice, resuspended in lysis buffer, vortexed, and then placed on ice. ATP was measured by luminometric methods using commercially available luciferin/luciferase reagents per the manufacturer’s instructions (ATP Assay Kit; Beyotime Biotech) in a luminometer (TD-20/20, Turner Designs). An average of 5 × 107 sperm was used for ATP analysis.
Supplementary Material
Acknowledgments
We thank Ryan M. Matzuk for performing the initial bioinformatic analysis that identified the Tcte1 gene; Jinyang Cai for microscopy; Wentao Zeng for zygote injections and transplantation; and Dr. John Nelson for critical review of the manuscript. This work was supported by the National Key Research and Development Program of China (2016YFA0500903); the National Basic Research Program of China (2015CB943003); the Natural Science Foundation of China (31530047 and 31571536); Eunice Kennedy Shriver National Institute of Child Health and Human Development Grant R01 HD088412 (to M.M.M. and M.I.); the Osaka University International Joint Research Promotion Program (to M.M.M. and M.I.); Ministry of Education, Culture, Sports, Science, and Technology KAKENHI Grant JP25112007 (to M.I.); the Takeda Science Foundation Grant (to H.M. and M.I); Baylor College of Medicine Training Grant 5T32HD007165-35 (to J.M.C.); the Academy of Finland and the Sigrid Juselius Foundation (to R.P.-H.); Wellcome Trust Grants 079643 and 098051 (to R.R.-S.); and National Institutes of Health [Knockout Mouse Project (KOMP)] Award U01-HG004080 (to R.R.-S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this article have been deposited in the ProteomeXchange Consortium with identifier PXD005343.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621279114/-/DCSupplemental.
References
- 1.Blocker A, Komoriya K, Aizawa S. Type III secretion systems and bacterial flagella: Insights into their function from structural similarities. Proc Natl Acad Sci USA. 2003;100:3027–3030. doi: 10.1073/pnas.0535335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. Evolution: Tracing the origins of centrioles, cilia, and flagella. J Cell Biol. 2011;194:165–175. doi: 10.1083/jcb.201011152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faguy DM, Jarrell KF, Kuzio J, Kalmokoff ML. Molecular analysis of archael flagellins: Similarity to the type IV pilin-transport superfamily widespread in bacteria. Can J Microbiol. 1994;40:67–71. doi: 10.1139/m94-011. [DOI] [PubMed] [Google Scholar]
- 4.Lindemann CB, Lesich KA. Functional anatomy of the mammalian sperm flagellum. Cytoskeleton. 2016;73:652–669. doi: 10.1002/cm.21338. [DOI] [PubMed] [Google Scholar]
- 5.Allen RD. A reinvestigation of cross-sections of cilia. J Cell Biol. 1968;37:825–831. doi: 10.1083/jcb.37.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibbons IR, Rowe AJ. Dynein: A Protein with adenosine triphosphatase activity from cilia. Science. 1965;149:424–426. doi: 10.1126/science.149.3682.424. [DOI] [PubMed] [Google Scholar]
- 7.Bower R, et al. The N-DRC forms a conserved biochemical complex that maintains outer doublet alignment and limits microtubule sliding in motile axonemes. Mol Biol Cell. 2013;24:1134–1152. doi: 10.1091/mbc.E12-11-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curi SM, et al. Asthenozoospermia: Analysis of a large population. Arch Androl. 2003;49:343–349. doi: 10.1080/01485010390219656. [DOI] [PubMed] [Google Scholar]
- 9.Yatsenko AN, Iwamori N, Iwamori T, Matzuk MM. The power of mouse genetics to study spermatogenesis. J Androl. 2010;31:34–44. doi: 10.2164/jandrol.109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matzuk MM, Lamb DJ. The biology of infertility: Research advances and clinical challenges. Nat Med. 2008;14:1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miki K, et al. Targeted disruption of the Akap4 gene causes defects in sperm flagellum and motility. Dev Biol. 2002;248:331–342. doi: 10.1006/dbio.2002.0728. [DOI] [PubMed] [Google Scholar]
- 12.Roy A, Lin YN, Agno JE, DeMayo FJ, Matzuk MM. Tektin 3 is required for progressive sperm motility in mice. Mol Reprod Dev. 2009;76:453–459. doi: 10.1002/mrd.20957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy A, Lin YN, Agno JE, DeMayo FJ, Matzuk MM. Absence of tektin 4 causes asthenozoospermia and subfertility in male mice. FASEB J. 2007;21:1013–1025. doi: 10.1096/fj.06-7035com. [DOI] [PubMed] [Google Scholar]
- 14.Young SA, et al. CABYR is essential for fibrous sheath integrity and progressive motility in mouse spermatozoa. J Cell Sci. 2016;129:4379–4387. doi: 10.1242/jcs.193151. [DOI] [PubMed] [Google Scholar]
- 15.Carlson AE, et al. CatSper1 required for evoked Ca2+ entry and control of flagellar function in sperm. Proc Natl Acad Sci USA. 2003;100:14864–14868. doi: 10.1073/pnas.2536658100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danshina PV, et al. Phosphoglycerate kinase 2 (PGK2) is essential for sperm function and male fertility in mice. Biol Reprod. 2010;82:136–145. doi: 10.1095/biolreprod.109.079699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miki K, et al. Glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme, is required for sperm motility and male fertility. Proc Natl Acad Sci USA. 2004;101:16501–16506. doi: 10.1073/pnas.0407708101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Odet F, et al. Expression of the gene for mouse lactate dehydrogenase C (Ldhc) is required for male fertility. Biol Reprod. 2008;79:26–34. doi: 10.1095/biolreprod.108.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avenarius MR, et al. Human male infertility caused by mutations in the CATSPER1 channel protein. Am J Hum Genet. 2009;84:505–510. doi: 10.1016/j.ajhg.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben Khelifa M, et al. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum Genet. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuccarello D, et al. Mutations in dynein genes in patients affected by isolated non-syndromic asthenozoospermia. Hum Reprod. 2008;23:1957–1962. doi: 10.1093/humrep/den193. [DOI] [PubMed] [Google Scholar]
- 22.Zuccarello D, et al. A possible association of a human tektin-t gene mutation (A229V) with isolated non-syndromic asthenozoospermia: Case report. Hum Reprod. 2008;23:996–1001. doi: 10.1093/humrep/dem400. [DOI] [PubMed] [Google Scholar]
- 23.Lin J, et al. Building blocks of the nexin-dynein regulatory complex in Chlamydomonas flagella. J Biol Chem. 2011;286:29175–29191. doi: 10.1074/jbc.M111.241760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adl SM, et al. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adl SM, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 26.Nelson GA, Roberts TM, Ward S. Caenorhabditis elegans spermatozoan locomotion: Amoeboid movement with almost no actin. J Cell Biol. 1982;92:121–131. doi: 10.1083/jcb.92.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 28.Miyata H, et al. Genome engineering uncovers 54 evolutionarily conserved and testis-enriched genes that are not required for male fertility in mice. Proc Natl Acad Sci USA. 2016;113:7704–7710. doi: 10.1073/pnas.1608458113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradley A, et al. The mammalian gene function resource: The International Knockout Mouse Consortium. Mamm Genome. 2012;23:580–586. doi: 10.1007/s00335-012-9422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyata H, et al. Sperm calcineurin inhibition prevents mouse fertility with implications for male contraceptive. Science. 2015;350:442–445. doi: 10.1126/science.aad0836. [DOI] [PubMed] [Google Scholar]
- 31.Sarvetnick N, Tsai JY, Fox H, Pilder SH, Silver LM. A mouse chromosome 17 gene encodes a testes-specific transcript with unusual properties. Immunogenetics. 1989;30:34–41. doi: 10.1007/BF02421467. [DOI] [PubMed] [Google Scholar]
- 32.Heuser T, Raytchev M, Krell J, Porter ME, Nicastro D. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J Cell Biol. 2009;187:921–933. doi: 10.1083/jcb.200908067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang B, Ramanis Z, Luck DJ. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell. 1982;28:115–124. doi: 10.1016/0092-8674(82)90381-6. [DOI] [PubMed] [Google Scholar]
- 34.Stephens RE. Isolation of nexin: The linkage protein responsible for maintenance of nine-fold configuration of flagellar axonemes. Biol Bull. 1970;139:438. [Google Scholar]
- 35.Satir P. Studies on cilia. 3. Further studies on the cilium tip and a “sliding filament” model of ciliary motility. J Cell Biol. 1968;39:77–94. doi: 10.1083/jcb.39.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wirschell M, et al. The nexin-dynein regulatory complex subunit DRC1 is essential for motile cilia function in algae and humans. Nat Genet. 2013;45:262–268. doi: 10.1038/ng.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oda T, Yanagisawa H, Kikkawa M. Detailed structural and biochemical characterization of the nexin-dynein regulatory complex. Mol Biol Cell. 2015;26:294–304. doi: 10.1091/mbc.E14-09-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakanishi T, Ikawa M, Yamada S, Toshimori K, Okabe M. Alkalinization of acrosome measured by GFP as a pH indicator and its relation to sperm capacitation. Dev Biol. 2001;237:222–231. doi: 10.1006/dbio.2001.0353. [DOI] [PubMed] [Google Scholar]
- 39.Fujihara Y, Kaseda K, Inoue N, Ikawa M, Okabe M. Production of mouse pups from germline transmission-failed knockout chimeras. Transgenic Res. 2013;22:195–200. doi: 10.1007/s11248-012-9635-x. [DOI] [PubMed] [Google Scholar]
- 40.Cao W, Gerton GL, Moss SB. Proteomic profiling of accessory structures from the mouse sperm flagellum. Mol Cell Proteomics. 2006;5:801–810. doi: 10.1074/mcp.M500322-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Ha S, Lindsay AM, Timms AE, Beier DR. Mutations in Dnaaf1 and Lrrc48 cause hydrocephalus, laterality defects, and sinusitis in mice. G3 (Bethesda) 2016;6:2479–2487. doi: 10.1534/g3.116.030791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Curtiss NP, et al. Isolation and analysis of candidate myeloid tumor suppressor genes from a commonly deleted segment of 7q22. Genomics. 2005;85:600–607. doi: 10.1016/j.ygeno.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 43.Ma Q, et al. Molecular cloning and characterization of SRG-L, a novel mouse gene developmentally expressed in spermatogenic cells. Mol Reprod Dev. 2006;73:1075–1083. doi: 10.1002/mrd.20485. [DOI] [PubMed] [Google Scholar]
- 44.Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.San Agustin JT, Pazour GJ, Witman GB. Intraflagellar transport is essential for mammalian spermiogenesis but is absent in mature sperm. Mol Biol Cell. 2015;26:4358–4372. doi: 10.1091/mbc.E15-08-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gervais FG, et al. Recruitment and activation of caspase-8 by the Huntingtin-interacting protein Hip-1 and a novel mpartner Hippi. Nat Cell Biol. 2002;4:95–105. doi: 10.1038/ncb735. [DOI] [PubMed] [Google Scholar]
- 47.Zhang B, Kirov S, Snoddy J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013. Nucleic Acids Res. 2013;41:W77–83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakamura N, Mori C, Eddy EM. Molecular complex of three testis-specific isozymes associated with the mouse sperm fibrous sheath: Hexokinase 1, phosphofructokinase M, and glutathione S-transferase mu class 5. Biol Reprod. 2010;82:504–515. doi: 10.1095/biolreprod.109.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ijiri TW, et al. Male mice express spermatogenic cell-specific triosephosphate isomerase isozymes. Mol Reprod Dev. 2013;80:862–870. doi: 10.1002/mrd.22217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mukai C, Okuno M. Glycolysis plays a major role for adenosine triphosphate supplementation in mouse sperm flagellar movement. Biol Reprod. 2004;71:540–547. doi: 10.1095/biolreprod.103.026054. [DOI] [PubMed] [Google Scholar]
- 52.McCarrey JR, Thomas K. Human testis-specific PGK gene lacks introns and possesses characteristics of a processed gene. Nature. 1987;326:501–505. doi: 10.1038/326501a0. [DOI] [PubMed] [Google Scholar]
- 53.Storey BT, Kayne FJ. Energy metabolism of spermatozoa. V. The Embden-Myerhof pathway of glycolysis: Activities of pathway enzymes in hypotonically treated rabbit epididymal spermatozoa. Fertil Steril. 1975;26:1257–1265. [PubMed] [Google Scholar]
- 54.Mori C, Welch JE, Fulcher KD, O’Brien DA, Eddy EM. Unique hexokinase messenger ribonucleic acids lacking the porin-binding domain are developmentally expressed in mouse spermatogenic cells. Biol Reprod. 1993;49:191–203. doi: 10.1095/biolreprod49.2.191. [DOI] [PubMed] [Google Scholar]
- 55.Sakai I, Sharief FS, Li SS. Molecular cloning and nucleotide sequence of the cDNA for sperm-specific lactate dehydrogenase-C from mouse. Biochem J. 1987;242:619–622. doi: 10.1042/bj2420619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Welch JE, Schatte EC, O’Brien DA, Eddy EM. Expression of a glyceraldehyde 3-phosphate dehydrogenase gene specific to mouse spermatogenic cells. Biol Reprod. 1992;46:869–878. doi: 10.1095/biolreprod46.5.869. [DOI] [PubMed] [Google Scholar]
- 57.Krisfalusi M, Miki K, Magyar PL, O’Brien DA. Multiple glycolytic enzymes are tightly bound to the fibrous sheath of mouse spermatozoa. Biol Reprod. 2006;75:270–278. doi: 10.1095/biolreprod.105.049684. [DOI] [PubMed] [Google Scholar]
- 58.Porter ME, Sale WS. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J Cell Biol. 2000;151:F37–F42. doi: 10.1083/jcb.151.5.f37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ihara M, et al. Cortical organization by the septin cytoskeleton is essential for structural and mechanical integrity of mammalian spermatozoa. Dev Cell. 2005;8:343–352. doi: 10.1016/j.devcel.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 60.Lhuillier P, et al. Absence of annulus in human asthenozoospermia: Case report. Hum Reprod. 2009;24:1296–1303. doi: 10.1093/humrep/dep020. [DOI] [PubMed] [Google Scholar]
- 61.Oji A, et al. CRISPR/Cas9 mediated genome editing in ES cells and its application for chimeric analysis in mice. Sci Rep. 2016;6:31666. doi: 10.1038/srep31666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen B, et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23:720–723. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang M, et al. Lack of testicular seipin causes teratozoospermia syndrome in men. Proc Natl Acad Sci USA. 2014;111:7054–7059. doi: 10.1073/pnas.1324025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tokuhiro K, et al. Meichroacidin containing the membrane occupation and recognition nexus motif is essential for spermatozoa morphogenesis. J Biol Chem. 2008;283:19039–19048. doi: 10.1074/jbc.M708590200. [DOI] [PubMed] [Google Scholar]
- 65.Fujihara Y, et al. SPACA1-deficient male mice are infertile with abnormally shaped sperm heads reminiscent of globozoospermia. Development. 2012;139:3583–3589. doi: 10.1242/dev.081778. [DOI] [PubMed] [Google Scholar]
- 66.Ikawa M, et al. Calsperin is a testis-specific chaperone required for sperm fertility. J Biol Chem. 2011;286:5639–5646. doi: 10.1074/jbc.M110.140152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Castañeda J, et al. Reduced pachytene piRNAs and translation underlie spermiogenic arrest in Maelstrom mutant mice. EMBO J. 2014;33:1999–2019. doi: 10.15252/embj.201386855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Osuru HP, et al. The acrosomal protein SP-10 (Acrv1) is an ideal marker for staging of the cycle of seminiferous epithelium in the mouse. Mol Reprod Dev. 2014;81:896–907. doi: 10.1002/mrd.22358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Villén J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hao P, Ren Y, Dutta B, Sze SK. Comparative evaluation of electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) and high-pH reversed phase (Hp-RP) chromatography in profiling of rat kidney proteome. J Proteomics. 2013;82:254–262. doi: 10.1016/j.jprot.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 71.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 72.Deutsch EW, et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vizcaíno JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu M, et al. Scanning of novel cancer/testis proteins by human testis proteomic analysis. Proteomics. 2013;13:1200–1210. doi: 10.1002/pmic.201200489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.