Fig. 1.
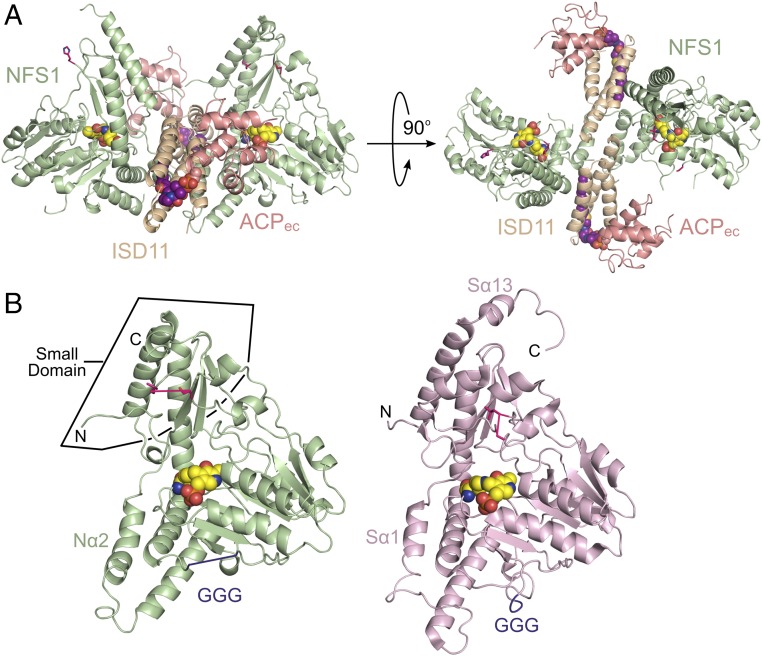
The architecture of a mitochondrial cysteine desulfurase. (A) Ribbon diagram of the SDAec structural architecture displayed with NFS1 (light green), ISD11 (wheat), and ACPec (salmon) in two orthogonal orientations. The K258-PLP (yellow) and lipid-bound 4’-PPT (magenta) cofactors are shown as spheres. (B) NFS1 and IscS (light pink; PDB ID code 3LVM) subunits displayed in similar orientations. Residues connecting disordered mobile S loop are shown in hot pink, whereas those connecting the GGG loop are in plum. The rmsd for 277 NFS1 (chain A) and IscS (chain A) Cα atoms is 0.84 Å.