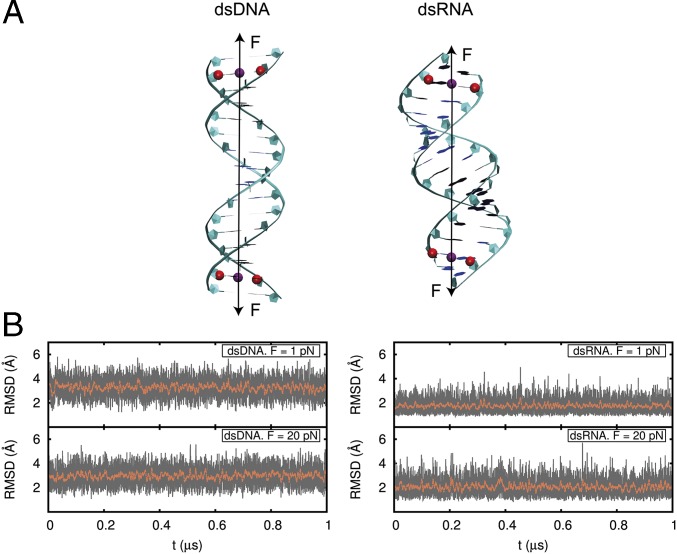
Fig. 1.
dsDNA and dsRNA molecules under a constant stretching force. (A) dsDNA and dsRNA molecule starting configurations were B-form DNA and A-form RNA. For both molecules, the force (black arrow) was implemented to act on the centers of mass of the C1′ atoms of the second and 15th base pairs (red). Five simulations of t ≥ 1 μs each were run for each molecule at force moduli of 1, 5, 10, 15, and 20 pN. (B) Computed rmsd values for the heavy atoms of the 10 central base pairs of dsDNA and dsRNA with respect to their standard B- and A-forms at every frame (1,000 steps of 2 fs) of the simulation (gray) and averaged over a running window of 2,000 frames (red).