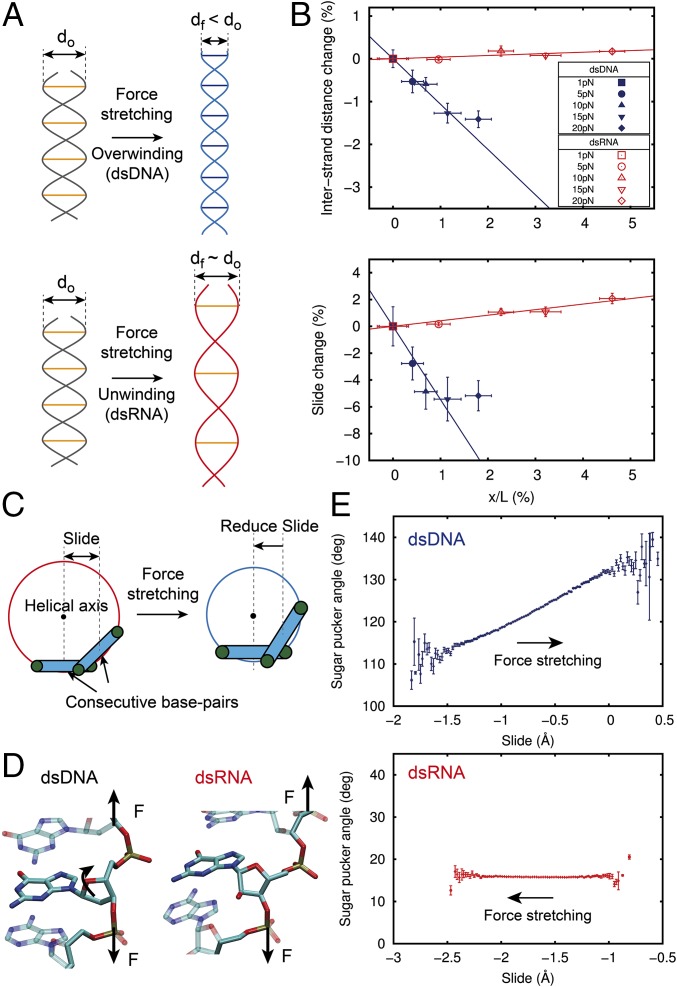
Fig. 4.
Physical mechanism of the opposite sign of dsDNA and dsRNA twist-stretch coupling. (A, Upper) Double helical structure can overwind when stretched if the interstrand distance is allowed to shrink. (A, Lower) Alternatively, a fixed interstrand distance duplex will unwind when stretched. (B) Relative change of the interstrand distance (Upper) and slide (Lower) with respect to the F = 1 pN value, plotted against the relative increase in the extension induced by force (dsDNA, blue; dsRNA, red). Datasets were fitted to a linear function constrained to pass through the (0,0) point, excluding the value at F = 20 pN for dsDNA (main text). Error bars were calculated as described in Materials and Methods. (C) Cartoon illustrating the relationship between slide and interstrand distance upon stretching. A reduction of slide is accompanied by a reduction of the interstrand distance as it occurs with dsDNA (SI Appendix, Fig. S8). (D) Representation of two base-pair steps to highlight the different orientation of the sugar with respect to the phosphate backbone of dsDNA (Left) and dsRNA (Right). (E) Fluctuations in sugar pucker angle with respect to the slide parameter. The bin size is 0.02 Å. Data points are mean values with SEM.