Abstract
The genus Phyllosticta occurs worldwide, and contains numerous plant pathogenic, endophytic and saprobic species. Phyllosticta citricarpa is the causal agent of Citrus Black Spot disease (CBS), affecting fruits and leaves of several citrus hosts (Rutaceae), and can also be isolated from asymptomatic citrus tissues. Citrus Black Spot occurs in citrus-growing regions with warm summer rainfall climates, but is absent in countries of the European Union (EU). Phyllosticta capitalensis is morphologically similar to P. citricarpa, but is a non-pathogenic endophyte, commonly isolated from citrus leaves and fruits and a wide range of other hosts, and is known to occur in Europe. To determine which Phyllosticta spp. occur within citrus growing regions of EU countries, several surveys were conducted (2015–2017) in the major citrus production areas of Greece, Italy, Malta, Portugal and Spain to collect both living plant material and leaf litter in commercial nurseries, orchards, gardens, backyards and plant collections. A total of 64 Phyllosticta isolates were obtained from citrus in Europe, of which 52 were included in a multi-locus (ITS, actA, tef1, gapdh, LSU and rpb2 genes) DNA dataset. Two isolates from Florida (USA), three isolates from China, and several reference strains from Australia, South Africa and South America were included in the overall 99 isolate dataset. Based on the data obtained, two known species were identified, namely P. capitalensis (from asymptomatic living leaves of Citrus spp.) in Greece, Italy, Malta, Portugal and Spain, and P. citricarpa (from leaf litter of C. sinensis and C. limon) in Italy, Malta and Portugal. Moreover, two new species were described, namely P. paracapitalensis (from asymptomatic living leaves of Citrus spp.) in Italy and Spain, and P. paracitricarpa (from leaf litter of C. limon) in Greece. On a genotypic level, isolates of P. citricarpa populations from Italy and Malta (MAT1-2-1) represented a single clone, and those from Portugal (MAT1-1-1) another. Isolates of P. citricarpa and P. paracitricarpa were able to induce atypical lesions (necrosis) in artificially inoculated mature sweet orange fruit, while P. capitalensis and P. paracapitalensis induced no lesions. The Phyllosticta species recovered were not found to be widespread, and were not associated with disease symptoms, indicating that the fungi persisted over time, but did not cause disease.
Key words: Citrus, Guignardia, Multi-locus sequence typing, Systematics
Taxonomic novelties: Phyllosticta paracapitalensis Guarnaccia & Crous, sp. nov.; P. paracitricarpa Guarnaccia & Crous, sp. nov
Introduction
The genus Phyllosticta was introduced by Persoon (1818), with P. convallariae (nom. cons.) (= P. cruenta) designated as the type species (Donk 1968). Species of Phyllosticta are known as plant pathogens of several hosts and responsible for various disease symptoms including leaf and fruit spots. Species included in the P. ampelicida species complex, which cause black rot disease on grapevines (Wicht et al., 2012, Zhou et al., 2015), and in the P. musarum species complex, which cause banana freckle disease, are economically important plant pathogens (Pu et al., 2008, Wong et al., 2012). Some species of Phyllosticta have also been isolated as endophytes from a wide range of hosts (e.g., P. capitalensis) and as saprobes (Glienke-Blanco et al., 2002, Huang et al., 2008, Thongkantha et al., 2008, Wikee et al., 2011, Wikee et al., 2013b).
Sexual morphs have in the past been named in Guignardia (van der Aa 1973). The name Guignardia was introduced as a replacement for Laestadia by Viala & Ravaz (1892), who applied the name only to Sphaeria bidwellii (= G. bidwellii = P. ampelicida), a species that is different from Laestadia (Bissett 1986). Petrak (1957) included G. bidwellii and related species in Botryosphaeria, an opinion that was initially shared by Barr, 1970, Barr, 1972. Phyllosticta was first monographed by van der Aa (1973) and more recently all species names described in Phyllosticta were re-described by van der Aa & Vanev (2002). Schoch et al. (2006) placed Phyllosticta in Botryosphaeriales. Several authors showed that Botryosphaeriaceae contained both Botryosphaeria and Phyllosticta spp., although this relationship remained poorly resolved (Crous et al., 2006, Schoch et al., 2006, Liu et al., 2012).
With the increasing use of molecular data to link asexual and sexual morphs, and the end of dual nomenclature for fungi (Hawksworth et al. 2011, Wingfield et al. 2012), the oldest and more commonly used name, Phyllosticta, was chosen over that of Guignardia (Glienke et al., 2011, Sultan et al., 2011, Wikee et al., 2011, Wikee et al., 2013b, Wong et al., 2012). Moreover, several studies incorporated DNA sequence data to improve the identification and help resolve the taxonomy of Phyllosticta spp. (Baayen et al., 2002, Wulandari et al., 2009, Glienke et al., 2011, Wikee et al., 2011). Presently, new species of Phyllosticta are described based on a polyphasic approach, incorporating phylogenetic data, morphology and culture characteristics (Crous et al., 2012, Su and Cai, 2012, Wang et al., 2012, Wong et al., 2012, Zhang et al. 2015). Wikee et al. (2013a) redefined Phyllosticta, showing that it clusters sister to the Botryosphaeriaceae for which the authors resurrected the family name Phyllostictaceae.
The main morphological characters used to recognise a species of Phyllosticta is the production of pycnidia containing aseptate, hyaline conidia that are covered by a mucoid layer and bearing a single apical appendage (van der Aa 1973). However, the mucoid layer and appendage are not always present. The sexual morph has erumpent, globose to pyriform ascomata, often irregularly shaped, unilocular, and with a central ostiole. Asci are 8-spored, bitunicate, clavate to broadly ellipsoid, with a wide, obtusely rounded or slightly square apex. Ascospores are ellipsoid to limoniform, sometimes slightly elongated, aseptate, hyaline, often with a large central guttule and a mucoid cap at each end. Spermatia produced in culture are hyaline, aseptate, cylindrical to dumbbell-shaped with guttules at both ends (van der Aa 1973).
Several Phyllosticta species have been associated with Citrus spp. worldwide (Baayen et al., 2002, Glienke-Blanco et al., 2002, Everett and Rees-George, 2006, Baldassari et al., 2008, Wulandari et al., 2009, Glienke et al., 2011, Brentu et al., 2012, Wikee et al., 2013a, Er et al., 2014). Citrus black spot (CBS) is a foliar and fruit disease of Citrus spp. caused by P. citricarpa (sexual morph Guignardia citricarpa) (Kotzé, 1981, Baldassari et al., 2008). The pathogen affects fruits and leaves of several citrus hosts causing various symptoms (Kiely, 1948a, Kiely, 1949, Kotzé, 1981, Kotzé, 2000, Snowdon, 1990) with lemons and ‘Valencia’ oranges being more susceptible (Kotzé 2000). Hard spot is the most common symptom characterised by sunken, pale brown necrotic lesions with a dark reddish brown raised border; lesions often containing the pycnidia (asexual sporocarps). Several other kinds of lesions are known: virulent spot, a sunken necrotic lesion without defined borders mostly on mature fruit; false melanose consisting of small black pustules usually in a tear stain pattern; and freckle, cracked or speckled spot. Leaf symptoms are seldom seen except on lemons. They appear as round, small, sunken necrotic spots with a yellow halo (Schubert et al. 2010). The infected leaves, when fallen on the orchard floor, represent a substrate for the development and maturation of pseudothecia from which the primary inoculum, ascospores, are released for new infections (McOnie 1967). Phyllosticta citricarpa has never been found on plant species outside of the Rutaceae, and can be isolated from asymptomatic citrus tissues (Baldassari et al. 2008).
Phyllosticta citricarpa is often associated with P. capitalensis, a morphologically similar but non-pathogenic species, previously incorrectly considered as the asexual morph of Guignardia mangiferae (Baayen et al., 2002, Everett and Rees-George, 2006, Glienke et al., 2011). Based on a multi-locus phylogenetic analysis, however, Glienke et al. (2011) revealed that P. capitalensis sensu lato was genetically distinct from the reference isolate of G. mangiferae. Phyllosticta capitalensis was initially described on Stanhopea (Orchidaceae) from Brazil (Hennings 1908). Okane et al. (2001) attributed the name P. capitalensis to an endophytic species reported on ericaceous plants from Japan, and described the sexual morph as a new species, G. endophyllicola. Subsequently Baayen et al. (2002), based on DNA sequence data of the ITS nrDNA, considered a common endophytic species associated with several plants as morphologically similar to G. endophyllicola, but attributed this species to G. mangiferae, while the asexual morph was referred to as P. capitalensis. Phyllosticta capitalensis is a cosmopolitan fungus that has been reported from plants in 21 different families (Johnston, 1998, Rodrigues and Samuels, 1999, Okane et al., 2001, Baayen et al., 2002, Glienke-Blanco et al., 2002, Rodrigues et al., 2004, Everett and Rees-George, 2006, Meyer et al., 2006, Rakotoniriana et al., 2008, Yuan et al., 2009, Bezerra et al., 2012) and has been found on citrus associated with both CBS affected and asymptomatic plants (Baayen et al., 2002, Everett and Rees-George, 2006, Glienke et al., 2011). Guignardia mangiferae sensu stricto (not P. capitalensis) causes angular leaf spots on mango (Baldassari et al., 2008, Glienke et al., 2011).
The biology and ecology of P. capitalensis differs from that of P. citricarpa. Phyllosticta capitalensis is homothallic, whereas P. citricarpa is heterothallic (Zhang et al., 2015, Wang et al., 2016, Amorim et al., 2017). Phyllosticta capitalensis produces fertile pseudothecia on agar media and P. citricarpa produces them on leaf litter (Kiely 1948a). Moreover, P. capitalensis is an ubiquitous, cosmopolitan endophyte of woody plants (Baayen et al. 2002) and P. citricarpa is associated only with citrus plants (Glienke et al. 2011).
Significant progress in species differentiation was achieved with multi-locus phylogenetic analyses performed on a large number of Phyllosticta species, (Wulandari et al., 2009, Glienke et al., 2011, Wang et al., 2012). Using three partial DNA regions, Wulandari et al. (2009) revealed three Phyllosticta clades associated with citrus in Thailand, namely P. capitalensis, P. citricarpa and P. citriasiana. Wang et al. (2012) described one new species associated with citrus in China, namely P. citrichinaensis, and also distinguished two subclades within P. citricarpa. Sequencing four partial regions of DNA, Glienke et al. (2011) distinguished a new species, Phyllosticta citribraziliensis, associated with Citrus sp. in Brazil. Phyllosticta citriasiana causes Citrus Tan Spot disease on Citrus maxima in Asia (Wulandari et al. 2009). Phyllosticta citrichinaensis is a weak pathogen on various citrus species in Asia, and P. citribraziliensis is non-pathogenic endophyte on citrus in Brazil (Glienke et al., 2011, Wang et al., 2012). A recent study added a sixth Phyllosticta species associated with citrus, namely P. citrimaxima, which was isolated from Citrus Tan Spot on fruit of C. maxima in Thailand (Wikee et al. 2013a).
Based on sequences of the rDNA internal transcribed spacer (ITS) region, the P. citricarpa and P. capitalensis clades are clearly distinct, with each species showing low levels of intraspecific variation (Okane et al., 2003, Rodrigues et al., 2004). Phyllostica citricarpa and P. capitalensis have several morphological and physiological differences: colonies of P. citricarpa produce a yellow halo on oatmeal agar (OA), the growth rate is generally faster in P. capitalensis, conidia are coated with a thicker mucoid layer than observed in P. citricarpa, and there is a higher level of hydrolytic enzyme production in P. citricarpa than in P. capitalensis (Baayen et al., 2002, Glienke et al., 2011, Romão et al., 2011).
Windborne P. citricarpa ascospores produced in pseudothecia (ascocarps) and waterborne conidia produced in pycnidia may cause infection on citrus (Kiely, 1948a, Kotzé, 1963, Kotzé, 1996, Kotzé, 2000). The ascospores are considered the primary source of inoculum in the CBS disease cycle, while conidia may serve for short distance wash-down dispersal by rain (Kiely, 1948a, Whiteside, 1967, Sposito et al., 2011). Alternate wetting and sun drying of leaves and mild to warm temperature fluctuations are favourable conditions for maturation of pseudothecia and ascospore discharge (Kiely, 1948a, Lee and Huang, 1973, Truter, 2010, Fourie et al., 2013, Hu et al., 2014). Subsequently, infection is dependent on the presence of long periods of free surface water and suitable microclimatic conditions (Kiely, 1948a, Kiely, 1948b, Kiely, 1949, Kotzé, 1963, Kotzé, 1981, McOnie, 1967). Leaf litter colonised by P. citricarpa serves as the primary inoculum source. Thus leaf litter plays an important role and its removal or enhanced decomposition results in improved inoculum management (Bellotte et al., 2009, Truter, 2010, Sposito et al., 2011). Pseudothecia develop 40–180 d after leaf fall, releasing mature ascospores during rainfall that are dispersed by wind (Kotzé, 1963, McOnie, 1964, Huang and Chang, 1972, Reis et al., 2006, Fourie et al., 2013, Dummel et al., 2015). Fruits are susceptible for 4–5 mo after petal fall (Kiely, 1948b, Schutte et al., 2003, Schutte et al., 2012, Miles et al., 2004). Therefore, the onset of rain, ascospore release and fruit susceptibility period are strongly correlated in summer rainfall regions resulting in fruit infection (Kotzé, 1963, McOnie, 1964, McOnie, 1967, Whiteside, 1967). Following a long latent period, the onset of symptom expression on fruit coincides with fruit ripening (Kiely, 1948a, Whiteside, 1967, Kotzé, 1981, Spósito et al., 2008).
Phyllosticta citricarpa has been recorded in Australia since the late 19th century, causing CBS disease, specifically in coastal regions of New South Wales and Queensland (Benson, 1895, Kotzé, 1981, Miles et al., 2013), but not from the hot, dry inland citrus orchards, and not in the winter rainfall regions in Australia (Broadbent 1995). Phyllosticta citricarpa has also been recorded in summer rainfall citrus-growing regions in several areas: South America (Argentina, Brazil, Uruguay, Venezuela; Garran, 1996, Kotzé, 2000, European Union, 2000, Paul et al., 2005), Central America (West Indies; Calavan 1960), North America (Dewdney et al., 2012, Schubert et al., 2012, Zavala et al., 2014), Asia (Bhutan, China, India, Indonesia, Philippines, Taiwan; Brodrick, 1969, European Union, 1998, Kotzé, 2000, European Union, 2000) and Africa (Ghana, Kenya, Mozambique, Nigeria, South Africa, Swaziland, Zambia, Zimbabwe; Doidge, 1929, Kotzé, 1981, Kotzé, 2000, European Union, 1998, Baayen et al., 2002, Brentu et al., 2012). Several fruit and leaf diseases caused by different fungi such as Colletotrichum and Alternaria spp. (Vicent et al., 2007, Aiello et al., 2015) are present in the EU citrus-producing countries; however, the CBS disease has not been reported (Baker et al. 2014). In addition to the general phytosanitary regulations applicable to the import of citrus propagating plant material, the import of citrus fruit into the EU is subject to phytosanitary regulations relating to P. citricarpa (EC2000/29/EC, 2000).
Recent epidemiological studies (Paul et al., 2005, Yonow et al., 2013, Magarey et al., 2015) have shown that the climatic conditions in the citrus growing regions within the EU are unsuitable for establishment of P. citricarpa and development of CBS disease, with only small, restricted Mediterranean coastal areas where the climatic conditions have at most marginal potential suitability. Considering that citrus plants were moved from Asia, where CBS is endemic and also regarded as the centre of origin of citrus, to Northern Africa and other countries around the Mediterranean Sea by traders, as early as the 5th century BC (Ramón-Laca, 2003, Mabberley, 2004, Nicolosi, 2007), it would be expected that P. citricarpa and/or other Phyllosticta spp. may have been introduced to these citrus-growing countries along with the hosts, especially since infected plant material is regarded as the means of long-distance spread of this pathogen (Kiely, 1948b, Kotzé, 1981). Likewise, there is always the possibility of illegal movement of citrus plant propagating material. Therefore, the potential occurrence of Phyllosticta spp. was included in a broad survey of fungal citrus pathogens undertaken in citrus growing regions within EU countries (Guarnaccia et al., 2017, Sandoval-Denis et al., 2018). During 2015–2017, several surveys were conducted in the major citrus production areas of the EU and included the following: (i) surveys of both fresh plant material and leaf litter in commercial nurseries and citrus orchards, gardens, backyards and plant collections, (ii) cultivation of as many Phyllosticta isolates as possible from this material, (iii) subject isolates to DNA sequence analyses combined with morphological characterisation, (iv) compare these results with data from other phylogenetic studies on Phyllosticta, (v) identification of genotypes and mating types of the P. citricarpa isolates found in this study and, (vi) to evaluate potential pathogenicity of the Phyllosticta spp. isolated.
Materials and methods
Sampling and isolation
The initial surveys were carried out in 2015 and 2016 covering a total of 95 sites located in some of the main citrus-producing regions of Europe (Table 1). Evaluations were conducted by sampling approx. 25 fruits, 25 twig portions, 50 living leaves and 50 leaves from the litter layer from each Citrus host present in each site investigated. Samples were collected from Andalusia, Mallorca, Valencia (Spain), Apulia, Calabria, Sicily (Italy), Algarve (Portugal), Crete, Mesolongi, Nafplio (Greece), Gozo and La Valletta (Malta) areas. Investigated citrus species included Australasian lime (Citrus australasica), citranges (Citrus sinensis × Poncirus trifoliata), citrons (C. medica, C. medica var. sarcodactylis), kumquat (C. japonica), limequats (Citrus ×floridana), calamondin (×Citrofortunella microcarpa), mandarins (C. reticulata), tangelo (C. ×tangelo), oranges (C. ×aurantium, C. ×bergamia, C. ×sinensis), pummelo (C. maxima), grapefruit (C. paradisi), limes (C. ×aurantifolia, C. ×hystrix, C. ×latifolia) and lemons (C. ×limon). New surveys were performed during December 2016 and January 2017 at the sites where P. citricarpa and P. paracitricarpa were found during the initial surveys (Table 1) to confirm these findings and to assay the presence of symptoms on fruit, leaves and twigs.
Table 1.
Location and characteristics of the investigated sites.
| City (country) | GPS coordinates | Site | Plant age (years) | Condition3 |
|---|---|---|---|---|
| Acitrezza (Italy) | 37.561077, 15.161086 | Backyard | 20–30 | Cultivated |
| Agia (Greece) | 35.465979, 23.921240 | Orchard | 5–10 | Cultivated |
| Algemesi (Spain) | 39.207638, −0.449773 | Orchard | 5–10 | Cultivated |
| Algemesi (Spain) | 39.196895, −0.470823 | Orchard | 5–10 | Cultivated |
| Alginet (Spain) | 39.260069, −0.458032 | Orchard | 10–15 | Cultivated |
| Alginet (Spain) | 39.251407, −0.416424 | Orchard | 5–10 | Cultivated |
| Alhaurin El Grande (Spain) | 36.645374, −4.677086 | Orchard | 15–25 | Unkept |
| Alhaurin El Grande (Spain) | 36.664689, −4.698184 | Orchard | 15–25 | Cultivated |
| Alikianos (Greece) | 35.456657, 23.908632 | Orchard | 15–25 | Cultivated |
| Alikianos (Greece) | 35.462384, 23.904367 | Orchard | 10–15 | Unkept |
| Alikianos (Greece) | 35.446440, 23.919798 | Orchard | 10–15 | Unkept |
| Alikianos (Greece) | 35.466216, 23.945558 | Orchard | 10–15 | Cultivated |
| Almeria (Spain) | 36.834637, −2.402932 | Experimental orchard | 15–25 | Cultivated |
| Almeria (Spain) | 36.828832, −2.402892 | Experimental orchard | 15–25 | Cultivated |
| Alzira (Spain) | 39.156963, −0.490723 | Orchard | 10–15 | Cultivated |
| Amfilochia (Greece) | 38.961381, 21.171635 | Orchard | 10–15 | Cultivated |
| Argo (Greece) | 37.628645, 22.742179 | Orchard | 10–15 | Cultivated |
| Argo (Greece) | 37.655558, 22.739309 | Orchard | 10–15 | Cultivated |
| Argos (Greece) | 37.686587, 22.661719 | Orchard | 10–15 | Cultivated |
| Arta (Greece)1 | 39.161719, 20.929585 | Backyard | 30–40 | Unkept |
| Arta (Greece) | 39.155661, 20.903791 | Orchard | 15–25 | Cultivated |
| Arta (Greece) | 39.160465, 20.918257 | Orchard | 5–10 | Cultivated |
| Barcellona P.G. (Italy) | 38.110560, 15.136794 | Nursery | 1–3 | Cultivated |
| Brucoli (Italy) | 37.294823, 15.110518 | Orchard | 15–25 | Cultivated |
| Canicattì (Italy) | 37.358434, 13.840898 | Backyard | 20–30 | Cultivated |
| Carruba (Italy) | 37.684625, 15.190943 | Orchard | 15–25 | Unkept |
| Castellò (Spain) | 39.903922, −0.086197 | Orchard | 10–15 | Cultivated |
| Castellò (Spain) | 39.883861, −0.088225 | Orchard | 10–15 | Cultivated |
| Castellò (Spain) | 39.884013, −0.090945 | Orchard | 10–15 | Cultivated |
| Cefalù (Italy) | 38.029481, 14.012267 | Backyard | 20–30 | Unkept |
| Chania (Greece) | 35.493153, 24.051141 | Orchard | 10–15 | Cultivated |
| Chania (Greece) | 35.477894, 23.948060 | Orchard | 10–15 | Cultivated |
| Comiso (Italy) | 36.943980, 14.637159 | Orchard | 15–25 | Unkept |
| Conceicao (Portugal) | 37.048481, −7.916927 | Orchard | 15–25 | Cultivated |
| Curiglia (Italy) | 38.767729, 16.203763 | Orchard | 20–30 | Unkept |
| El Ejido (Spain) | 36.795207, −2.719992 | Orchard | 20–30 | Cultivated |
| Estellencs (Spain) | 39.653504, 2.481876 | Backyard | 30–40 | Unkept |
| Faro (Portugal) | 37.108457, −7.916805 | Orchard | 20–30 | Unkept |
| Faro (Portugal) | 37.062641, −7.917432 | Orchard | 10–15 | Unkept |
| Giarratana (Italy) | 36.883438, 14.974420 | Orchard | 10–15 | Cultivated |
| Gouria (Greece) | 38.454977, 21.257646 | Orchard | 15–25 | Cultivated |
| Gozo (Malta) | 36.049069, 14.259299 | Backyard | 20–30 | Unkept |
| Gozo (Malta) | 36.037531, 14.260120 | Orchard | 10–15 | Unkept |
| Gozo (Malta) | 36.049646, 14.279360 | Orchard | 15–25 | Cultivated |
| Gozo (Malta)2 | 36.055138, 14.259907 | Backyard | 60–70 | Unkept |
| Gozo (Malta) | 36.058166, 14.284453 | Backyard | 60–70 | Unkept |
| Grotte (Italy) | 37.679925, 15.177006 | Orchard | 15–25 | Cultivated |
| Guardia (Italy) | 37.662709, 15.175918 | Orchard | 15–25 | Cultivated |
| Kirtomados (Greece) | 35.478749, 23.916661 | Orchard | 15–25 | Cultivated |
| Leni (Italy) | 38.044422, 14.597517 | Backyard | 30–40 | Cultivated |
| Leni (Italy) | 38.552889, 14.827128 | Backyard | 30–40 | Cultivated |
| Lentini (Italy) | 37.320577, 15.020901 | Orchard | 15–25 | Cultivated |
| Malaga (Spain) | 36.761761, −4.427060 | Botanical garden | 40–50 | Unkept |
| Mascali (Italy) | 37.767684, 15.192503 | Nursery | 1–3 | Cultivated |
| Mascali (Italy) | 37.768258, 15.194639 | Nursery | 1–3 | Cultivated |
| Massafra (Italy) | 40.544756, 17.144112 | Orchard | 10–15 | Cultivated |
| Mastro (Greece) | 38.430287, 21.280539 | Orchard | 15–25 | Cultivated |
| Mesquita (Portugal) | 37.213673, −8.289493 | Orchard | 10–15 | Cultivated |
| Mesquita (Portugal) | 37.204525, −8.297812 | Orchard | 20–30 | Unkept |
| Mineo (Italy) | 37.350719, 14.690858 | Orchard | 15–25 | Cultivated |
| Moncada (Spain) | 39.588547, −0.394583 | Experimental orchard | 10–15 | Cultivated |
| Monchique (Portugal) | 37.332409, −8.514506 | Backyard | 20–30 | Unkept |
| Monchique (Portugal) | 37.336226, −8.503686 | Backyard | 20–30 | Unkept |
| Monchique (Portugal) | 37.332239, −8.492232 | Backyard | 20–30 | Unkept |
| Monchique (Portugal)2 | 37.326195, −8.526232 | Backyard | 30–40 | Unkept |
| Motta S. Anastasia (Italy) | 37.482099, 14.886016 | Orchard | 15–25 | Cultivated |
| Motta S. Anastasia (Italy) | 37.469713, 14.954161 | Orchard | 15–25 | Cultivated |
| Nafplio (Greece) | 37.589312, 22.785267 | Orchard | 10–15 | Unkept |
| Nafplio (Greece) | 37.575095, 22.695589 | Orchard | 15–25 | Cultivated |
| Nafplio (Greece) | 37.582292, 22.696803 | Orchard | 10–15 | Cultivated |
| Nafplio (Greece) | 37.588798, 22.874844 | Backyard | 10–15 | Cultivated |
| Nicolosi (Italy) | 37.611273, 15.029477 | Backyard | 5–10 | Cultivated |
| Niscemi (Italy) | 37.139783, 14.393402 | Backyard | 15–25 | Cultivated |
| Noto (Italy) | 36.846497, 15.095445 | Orchard | 15–25 | Unkept |
| Pachino (Italy) | 36.720032, 15.086993 | Backyard | 15–25 | Unkept |
| Pachino (Italy) | 36.722328, 15.089408 | Orchard | 15–25 | Unkept |
| Pedara (Italy) | 37.608708, 15.066544 | Backyard | 30–40 | Cultivated |
| Pizzo Calabro (Italy) | 38.760390, 16.226005 | Orchard | 15–25 | Cultivated |
| Ribera (Italy) | 37.497113, 13.241850 | Orchard | 5–10 | Cultivated |
| Ribera (Italy) | 37.504423, 13.252070 | Orchard | 5–10 | Cultivated |
| Riposto (Italy) | 37.696470, 15.199345 | Nursery | 1–3 | Cultivated |
| Rocca Imperiale (Italy) | 40.108385, 16.617951 | Orchard | 10–15 | Cultivated |
| San Gregorio (Italy) | 37.562297, 15.100965 | Backyard | 60–70 | Cultivated |
| Scordia (Italy) | 37.281526, 14.869149 | Orchard | 15–25 | Cultivated |
| Seville (Spain) | 37.508538, −5.962815 | Orchard | 15–25 | Cultivated |
| Seville (Spain) | 37.482978, −5.954910 | Orchard | 15–25 | Unkept |
| Sikoula (Greece) | 39.085933, 21.083398 | Orchard | 10–15 | Cultivated |
| Silves (Portugal) | 37.164148, −8.390841 | Orchard | 15–25 | Unkept |
| Soller (Spain) | 39.764529, 2.709609 | Botanical garden | 30–40 | Cultivated |
| Soller (Spain) | 39.770115, 2.726600 | Orchard | 20–30 | Cultivated |
| Terme Vigliatore (Italy) | 38.145801, 15.163235 | Nursery | 1–3 | Cultivated |
| Torremolinos (Spain) | 36.672722, −4.504134 | Orchard | 30–40 | Cultivated |
| Trebisacce (Italy)2 | 39.910122, 16.564824 | Backyard | 20–30 | Cultivated |
| Trebisacce (Italy) | 39.906711, 16.560634 | Orchard | 3–6 | Cultivated |
| Zurrieq (Malta)2 | 35.823845, 14.505099 | Backyard | 15–25 | Unkept |
Site where P. paracitricarpa isolates were found associated with leaf litter sampled.
Sites where P. citricarpa isolates were found associated with leaf litter sampled.
Cultivated: Plants kept under constant agronomical management. Unkept: Plants abandoned.
Fungal isolates were obtained using two procedures. In the first, leaf and fruit sections (5 × 5 mm) were aseptically cut and surface-sterilised in a sodium hypochlorite solution (10 %) for 20 s, followed by 70 % ethanol for 30 s, and rinsed three times in autoclaved water. The sections were dried on autoclaved tissue paper, placed on malt extract agar (MEA; Crous et al. 2009) amended with 100 μg/mL penicillin and 100 μg/mL streptomycin (MEA-PS) and incubated at 25 °C until characteristic Phyllosticta colonies were observed. In the second procedure, leaf litter, living leaves, fruits and twig portions were incubated in moist chambers at room temperature (25 °C ± 3 °C) for up to 14 d and inspected daily for fungal sporulation. Sporulating pycnidia obtained through both procedures were collected and crushed in a drop of sterile water and then spread over the surface of MEA-PS plates. After 24–36 h germinating spores were individually transferred onto MEA plates. The isolates used in this study are maintained in the Westerdijk Fungal Biodiversity Institute (CBS culture collection), Utrecht, The Netherlands, and in the working collection of Pedro Crous (CPC), housed at the Westerdijk Institute. In addition, two Phyllosticta isolates collected in Florida, USA (CPC 25312, CPC 25327) and three from China (ZJUCC200933, ZJUCC200937, ZJUCC200952) were included in the phylogenetic analyses. Sequences from additional species were retrieved from NCBI's GenBank nucleotide database. A total of 111 Phyllosticta (incl. 64 European) isolates were included in the study (Table 2), of which 100 (incl. the outgroup, Neofusicoccum mediterraneum CBS 121718) were used in the phylogenetic analysis.
Table 2.
Collection details and GenBank accession numbers of isolates included in this study.
| Species | Culture no.1 | Host | Country | Mating type idiomorph | GenBank no.2 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| ITS | actA | tef1 | gapdh | LSU | rpb2 | |||||
| Neofusicoccum mediterraneum | CBS 121718 | Eucalyptus sp. | Greece | – | GU251176 | KY855639 | GU251308 | KY855694 | KY855754 | KY855815 |
| Phyllosticta aloeicola | CPC 21020 = CBS 136058 | Aloe ferox | South Africa | – | KF154280 | KF289311 | KF289193 | KF289124 | KF206214 | KY855816 |
| CPC 21021 | Aloe ferox | South Africa | – | KF154281 | KF289312 | KF289194 | KF289125 | KF206213 | KY855817 | |
| P. bifrenariae | CBS 128855 = VIC30556 | Bifrenaria harrisoniae, leaf | Brazil | – | JF343565 | JF343649 | JF343586 | JF343744 | KF206209 | KY855818 |
| CPC 17467 | Bifrenaria harrisoniae, leaf | Brazil | – | KF170299 | KF289283 | KF289207 | KF289138 | KF206260 | KY855819 | |
| P. capitalensis | CBS 226.77 | Paphiopedilum callosum, leaf spot | Germany | – | FJ538336 | FJ538452 | FJ538394 | JF343718 | KF206289 | KY855820 |
| CBS 100175 | Citrus sp. | Brazil | – | FJ538320 | FJ538436 | FJ538378 | JF343699 | KF206327 | KY855821 | |
| CBS 101228 | Nephelium lappaceum | Hawaii | – | FJ538319 | FJ538435 | FJ538377 | KF289086 | KF206325 | KY855822 | |
| CBS 114751 | Vaccinium sp., leaf | New Zealand | – | FJ538349 | FJ538465 | FJ538407 | KF289088 | EU167584 | KY855823 | |
| CBS 123373 | Musa paradisiaca | Thailand | – | FJ538341 | FJ538457 | FJ538399 | JF343703 | JQ743604 | KY855824 | |
| CBS 123374 | Citrus aurantium | Thailand | – | FJ538332 | FJ538448 | FJ538390 | JF343702 | KY855755 | KY855825 | |
| CBS 128856 = CPC 18848 | Stanhopea sp. | Brazil | – | JF261465 | JF343647 | JF261507 | JF343776 | KF206304 | KY855826 | |
| CPC 14609 | Zyzygium sp. | Madagascar | – | KF206184 | KF289264 | KF289175 | KF289081 | KF206280 | KY855827 | |
| CPC 20259 | Orchidaceae | Thailand | – | KC291340 | KC342537 | KC342560 | KF289104 | KF206244 | KY855828 | |
| CPC 20263 | Magnoliaceae | Thailand | – | KC291341 | KC342538 | KC342561 | KF289085 | KF206241 | KY855829 | |
| CPC 20268 | Hibiscus syriacus | Thailand | – | KC291343 | KC342540 | KC342563 | KF289117 | KF206236 | KY855830 | |
| CPC 20275 | Polyalthia longifolia | Thailand | – | KC291347 | KC342544 | KC342567 | KF289107 | KF206230 | KY855831 | |
| CPC 20278 | Euphorbia milii | Thailand | – | KC291347 | KC342544 | KC342567 | KF289107 | KF206230 | KY855832 | |
| CPC 20508 | Ixora chinensis | Thailand | – | KF206198 | KF289302 | KF289185 | KF289111 | KF206225 | KY855833 | |
| CPC 25327 | Citrus sinensis | Florida | – | KY855585 | KY855640 | KY855914 | KY855695 | KY855756 | KY855834 | |
| CPC 27059 | Citrus limon, leaf | Italy | – | KY855586 | KY855641 | KY855915 | KY855696 | KY855757 | KY855835 | |
| CPC 27060 | Citrus limon, leaf | Italy | – | KY855587 | KY855642 | KY855916 | KY855697 | KY855758 | KY855836 | |
| CPC 27061 | Citrus limon, leaf | Italy | – | KY855588 | KY855643 | KY855917 | KY855698 | KY855759 | KY855837 | |
| CPC 27062 | Citrus limon, leaf | Italy | – | KY855589 | KY855644 | KY855918 | KY855699 | KY855760 | KY855838 | |
| CPC 27084 = CBS 141345 | Citrus aurantifolia, leaf | Italy | – | KY855590 | KY855645 | KY855919 | KY855700 | KY855761 | KY855839 | |
| CPC 27085 | Citrus aurantifolia, leaf | Italy | – | KY855591 | KY855646 | KY855920 | KY855701 | KY855762 | KY855840 | |
| CPC 27086 | Citrus aurantifolia, leaf | Italy | – | KY855592 | KY855647 | KY855921 | KY855702 | KY855763 | KY855841 | |
| CPC 27087 | Citrus aurantifolia, leaf | Italy | – | KY855593 | KY855648 | KY855922 | KY855703 | KY855764 | KY855842 | |
| CPC 27786 | Citrus limon, leaf | Greece | – | KY855594 | KY855649 | KY855923 | KY855704 | KY855765 | KY855843 | |
| CPC 27787 | Citrus limon, leaf | Greece | – | KY855595 | KY855650 | KY855924 | KY855705 | KY855766 | KY855844 | |
| CPC 27788 | Citrus limon, leaf | Greece | – | KY855596 | KY855651 | KY855925 | KY855706 | KY855767 | KY855845 | |
| CPC 27789 | Citrus limon, leaf | Greece | – | KY855597 | KY855652 | KY855926 | KY855707 | KY855768 | KY855846 | |
| CPC 27825 = CBS 141346 | C. medica var. sarcodactylis, leaf spot | Italy | – | KY855598 | KY855653 | KY855927 | KY855708 | KY855769 | KY855847 | |
| CPC 27826 | C. medica var. sarcodactylis, leaf spot | Italy | – | KY855599 | KY855654 | KY855928 | KY855709 | KY855770 | KY855848 | |
| CPC 27827 | C. medica var. sarcodactylis, leaf spot | Italy | – | KY855600 | KY855655 | KY855929 | KY855710 | KY855771 | KY855849 | |
| CPC 27828 | C. medica var. sarcodactylis, leaf spot | Italy | – | KY855601 | KY855656 | KY855930 | KY855711 | KY855772 | KY855850 | |
| CPC 27917 = CBS 141347 | Citrus limon, leaf | Malta | – | KY855602 | KY855657 | KY855931 | KY855712 | KY855773 | KY855851 | |
| CPC 27918 | Citrus limon, leaf | Malta | – | KY855603 | KY855658 | KY855932 | KY855713 | KY855774 | KY855852 | |
| CPC 27919 = CBS 141348 | Citrus limon, leaf | Portugal | – | KY855604 | KY855659 | KY855933 | KY855714 | KY855775 | KY855853 | |
| CPC 27920 | Citrus limon, leaf | Portugal | – | KY855605 | KY855660 | KY855934 | KY855715 | KY855776 | KY855854 | |
| CPC 28124 | Citrus limon, leaf | Spain | – | KY855606 | KY855661 | KY855935 | KY855716 | KY855777 | KY855855 | |
| CPC 28125 | Citrus limon, leaf | Spain | – | KY855607 | KY855662 | KY855936 | KY855717 | KY855778 | KY855856 | |
| CPC 28126 | Citrus limon, leaf | Spain | – | KY855608 | KY855663 | KY855937 | KY855718 | KY855779 | KY855857 | |
| P. citriasiana | CBS 120486 | Citrus maxima, fruit | Thailand | – | FJ538360 | FJ538476 | FJ538418 | JF343686 | KF206314 | KY855858 |
| CBS 120487 | Citrus maxima, fruit | China | – | FJ538361 | FJ538477 | FJ538419 | JF343687 | KF206313 | KY855859 | |
| CBS 123370 | Citrus maxima, fruit | Vietnam | – | FJ538355 | FJ538471 | FJ538413 | JF343689 | KF206310 | KY855860 | |
| P. citribraziliensis | CBS 100098 | Citrus sp., leaf | Brazil | – | FJ538352 | FJ538468 | FJ538410 | JF343691 | KF206221 | KY855861 |
| CPC 17464 | Citrus sp., leaf | Brazil | – | KF170300 | KF289280 | KF289224 | KF289159 | KF206263 | KY855862 | |
| CPC 17465 | Citrus sp., leaf | Brazil | – | KF170301 | KF289281 | KF289225 | KF289160 | KF206262 | KY855863 | |
| P. citricarpa | CBS 122482 | Citrus sinensis | Zimbabwe | MAT1-2-1 | FJ538317 | KF289265 | FJ538375 | KF289146 | KF306230 | KY855864 |
| CBS 127452 | Citrus reticulata | Australia | MAT1-2-1 | JF343581 | JF343665 | JF343602 | JF343769 | KF206307 | KY855865 | |
| CBS 127454 | Citrus limon | Australia | MAT1-2-1 | JF343583 | JF343667 | JF343604 | JF343771 | KF206306 | KY855866 | |
| CPC 16151 | Citrus sp. | South Africa | MAT1-1-1 | KF170291 | KF289267 | KF289221 | KF289156 | KF206276 | KY855867 | |
| CPC 16586 | Citrus limon | Argentina | MAT1-1-1 | KF170293 | KF289269 | KF289220 | KF289155 | KF206274 | KY855868 | |
| CPC 16603 | Citrus limon | Uruguay | MAT1-1-1 | KF170295 | KF289274 | KF289213 | KF289147 | KF206269 | KY855869 | |
| CPC 16609 | Citrus sp. | Argentina | MAT1-1-1 | KF170298 | KF289277 | KF289217 | KF289152 | KF206266 | KY855870 | |
| CPC 25312 | Citrus sinensis | Florida | MAT1-2-1 | KY855609 | KY855664 | KY855938 | KY855719 | KY855780 | KY855871 | |
| CPC 279093 = CBS 141349 | Citrus limon, leaf litter | Italy | MAT1-2-1 | KY855610 | KY855665 | KY855939 | KY855720 | KY855781 | KY855872 | |
| CPC 279103 | Citrus limon, leaf litter | Italy | MAT1-2-1 | KY855611 | KY855666 | KY855940 | KY855721 | KY855782 | KY855873 | |
| CPC 279113 | Citrus limon, leaf litter | Italy | MAT1-2-1 | KY855612 | KY855667 | KY855941 | KY855722 | KY855783 | KY855874 | |
| CPC 279123 | Citrus limon, leaf litter | Italy | MAT1-2-1 | KY855613 | KY855668 | KY855942 | KY855723 | KY855784 | KY855875 | |
| CPC 279133 = CBS 141350 | Citrus sinensis, leaf litter | Malta | MAT1-2-1 | KY855614 | KY855669 | KY855943 | KY855724 | KY855785 | KY855876 | |
| CPC 279143 | Citrus sinensis, leaf litter | Malta | MAT1-2-1 | KY855615 | KY855670 | KY855944 | KY855725 | KY855786 | KY855877 | |
| CPC 279153 | Citrus sinensis, leaf litter | Malta | MAT1-2-1 | KY855616 | KY855671 | KY855945 | KY855726 | KY855787 | KY855878 | |
| CPC 279163 | Citrus sinensis, leaf litter | Malta | MAT1-2-1 | KY855617 | KY855672 | KY855946 | KY855727 | KY855788 | KY855879 | |
| CPC 281043 = CBS 141351 | Citrus sinensis, leaf litter | Portugal | MAT1-1-1 | KY855618 | KY855673 | KY855947 | KY855728 | KY855789 | KY855880 | |
| CPC 281053 = CBS 141352 | Citrus sinensis, leaf litter | Portugal | MAT1-1-1 | KY855619 | KY855674 | KY855948 | KY855729 | KY855790 | KY855881 | |
| CPC 281063 | Citrus sinensis, leaf litter | Portugal | MAT1-1-1 | KY855620 | KY855675 | KY855949 | KY855730 | KY855791 | KY855882 | |
| CPC 281073 | Citrus sinensis, leaf litter | Portugal | MAT1-1-1 | KY855621 | KY855676 | KY855950 | KY855731 | KY855792 | KY855883 | |
| CPC 311713 | Citrus sinensis, leaf litter | Malta | MAT1-2-1 | – | – | – | – | – | – | |
| CPC 311723 | Citrus sinensis, leaf litter | Malta | MAT1-2-1 | – | – | – | – | – | – | |
| CPC 311733 | Citrus sinensis, leaf litter | Malta | MAT1-2-1 | – | – | – | – | – | – | |
| CPC 311743 | Citrus sinensis, leaf litter | Malta | MAT1-2-1 | – | – | – | – | – | – | |
| CPC 312793 | Citrus sinensis, leaf litter | Portugal | MAT1-1-1 | – | – | – | – | – | – | |
| CPC 312803 | Citrus sinensis, leaf litter | Portugal | MAT1-1-1 | – | – | – | – | – | – | |
| CPC 312813 | Citrus sinensis, leaf litter | Portugal | MAT1-1-1 | – | – | – | – | – | – | |
| CPC 312823 | Citrus sinensis, leaf litter | Portugal | MAT1-1-1 | – | – | – | – | – | – | |
| ZJUCC200952 | Citrus reticulata, leaf | China | MAT1-2-1 | JN791635 | JN791556 | JN791480 | KY855732 | KY855793 | KY855884 | |
| P. citrichinaensis | CBS 129764 = ZJUCC2010100 | Citrus reticulata, leaf | China | – | JN791598 | JN791527 | JN791453 | KY855733 | KY855794 | KY855885 |
| CBS 130529 = ZJUCC201085 = CGMCC3.14302 | Citrus maxima, leaf | China | – | JN791597 | JN791526 | JN791452 | KY855734 | KY855795 | KY855886 | |
| P. citrimaxima | CPC 20276 = CBS 136059 = MFLUCC10-0137 | Citrus maxima, fruit | Thailand | – | KF170304 | KF289300 | KF289222 | KF289157 | KF206229 | – |
| P. cordylinophila | CPC 20261 = MFLUCC10-0166 | Cordyline fruticosa | Thailand | – | KF170287 | KF289295 | KF289172 | KF289076 | KF206242 | KY855887 |
| CPC 20277 = MFLUCC12-0014 | Cordyline fruticosa | Thailand | – | KF170288 | KF289301 | KF289171 | KF289075 | KF206228 | KY855888 | |
| P. cussonia | CPC 14873 | Cussonia sp. | South Africa | – | JF343578 | JF343662 | JF343599 | JF343764 | KF206279 | KY855889 |
| CPC 14875 | Cussonia sp. | South Africa | – | JF343579 | JF343663 | JF343600 | JF343765 | KF206278 | KY855890 | |
| P. eugeniae | CBS 445.82 | Eugenia aromatica | Indonesia | – | AY042926 | KF289246 | KF289208 | KF289139 | KF206288 | KY855891 |
| P. hypoglossi | CBS 434.92 | Ruscus aculeatus | Italy | – | FJ538367 | FJ538483 | FJ538425 | JF343695 | KF206299 | KY855892 |
| P. paracapitalensis | CBS 173.77 | Citrus aurantiifolia | New Zealand | – | KF206179 | KF289244 | FJ538393 | KF289100 | KF306231 | KY855893 |
| CPC 26517 = CBS 141353 | Citrus floridana, leaf | Italy | – | KY855622 | KY855677 | KY855951 | KY855735 | KY855796 | KY855894 | |
| CPC 26518 | Citrus floridana, leaf | Italy | – | KY855623 | KY855678 | KY855952 | KY855736 | KY855797 | KY855895 | |
| CPC 26700 = CBS 141354 | Citrus floridana, leaf | Italy | – | KY855624 | KY855679 | KY855953 | KY855737 | KY855798 | KY855896 | |
| CPC 26701 | Citrus floridana, leaf | Italy | – | KY855625 | KY855680 | KY855954 | KY855738 | KY855799 | KY855897 | |
| CPC 26805 | Citrus floridana, leaf | Italy | – | KY855626 | KY855681 | KY855955 | KY855739 | KY855800 | KY855898 | |
| CPC 26806 | Citrus floridana, leaf | Italy | – | KY855627 | KY855682 | KY855956 | KY855740 | KY855801 | KY855899 | |
| CPC 28120 = CBS 141355 | Citrus limon, leaf | Spain | – | KY855628 | KY855683 | KY855957 | KY855741 | KY855802 | KY855900 | |
| CPC 28121 | Citrus limon, leaf | Spain | – | KY855629 | KY855684 | KY855958 | KY855742 | KY855803 | KY855901 | |
| CPC 28122 | Citrus limon, leaf | Spain | – | KY855630 | KY855685 | KY855959 | KY855743 | KY855804 | KY855902 | |
| CPC 28123 | Citrus limon, leaf | Spain | – | KY855631 | KY855686 | KY855960 | KY855744 | KY855805 | KY855903 | |
| CPC 28127 = CBS 141356 | Citrus limon, leaf | Spain | – | KY855632 | KY855687 | KY855961 | KY855745 | KY855806 | KY855904 | |
| CPC 28128 | Citrus limon, leaf | Spain | – | KY855633 | KY855688 | KY855962 | KY855746 | KY855807 | KY855905 | |
| CPC 28129 | Citrus limon, leaf | Spain | – | KY855634 | KY855689 | KY855963 | KY855747 | KY855808 | KY855906 | |
| P. paracitricarpa | CPC 27169 = CBS 141357 | Citrus limon, leaf litter | Greece | – | KY855635 | KY855690 | KY855964 | KY855748 | KY855809 | KY855907 |
| CPC 27170 = CBS 141358 | Citrus limon, leaf litter | Greece | – | KY855636 | KY855691 | KY855965 | KY855749 | KY855810 | KY855908 | |
| CPC 27171 = CBS 141359 | Citrus limon, leaf litter | Greece | – | KY855637 | KY855692 | KY855966 | KY855750 | KY855811 | KY855909 | |
| CPC 27172 = CBS 141360 | Citrus limon, leaf litter | Greece | – | KY855638 | KY855693 | KY855967 | KY855751 | KY855812 | KY855910 | |
| CPC 31246 | Citrus limon, leaf litter | Greece | – | – | – | – | – | – | – | |
| CPC 31247 | Citrus limon, leaf litter | Greece | – | – | – | – | – | – | – | |
| CPC 31248 | Citrus limon, leaf litter | Greece | – | – | – | – | – | – | – | |
| CPC 31249 | Citrus limon, leaf litter | Greece | – | – | – | – | – | – | – | |
| ZJUCC200933 | Citrus sinensis, fruit | China | – | JN791626 | JN791544 | JN791468 | KY855752 | KY855813 | KY855911 | |
| ZJUCC200937 | Citrus sinensis, fruit | China | – | JN791627 | JN791546 | JN791470 | KY855753 | KY855814 | KY855912 | |
| P. spinarum | CBS 292.90 | Chamaecyparis pisifera | France | – | JF343585 | JF343669 | JF343606 | JF343773 | KF206301 | KY855913 |
CPC: Culture collection of P.W. Crous, housed at CBS; CBS: CBS-KNAW Fungal Biodiversity Centre, Utrecht, the Netherlands; ZJUCC: Zhejiang University Culture Collection, China; MFLUCC: Mae Fah Luang University Culture Collection; CGMCC: China, General Microbiological Culture Collection, Beijing, China; VIC: Culture collection of Federal University of Viçosa, Viçosa, Brazil. Ex-type and ex-epitype cultures are indicated in bold.
ITS: internal transcribed spacers 1 and 2 together with 5.8S nrDNA; actA: partial actin gene; tef1: partial translation elongation factor 1-α gene; gapdh: partial glyceraldehyde-3-phosphate dehydrogenase gene; LSU: partial 28S (large subunit) nrDNA; rpb2: partial RNA polymerase II second largest subunit gene. Sequences generated in this study indicated in italics.
P. citricarpa isolates genotyped in this study.
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted using a Wizard® Genomic DNA Purification Kit (Promega Corporation, WI, USA) following the manufacturer's instructions. Partial regions of six loci were amplified. The primers V9G (de Hoog & Gerrits van den Ende 1998) and ITS4 (White et al. 1990) were used to amplify the internal transcribed spacer region (ITS) of the nuclear ribosomal RNA operon, including the 3′ end of the 18S rRNA, the first internal transcribed spacer region, the 5.8S rRNA gene; the second internal transcribed spacer region and the 5′ end of the 28S rRNA gene. The primers EF1-728F (Carbone & Kohn 1999) and EF2 (O’Donnell et al. 1998) were used to amplify part of the translation elongation factor 1-α gene (tef1). The primers ACT-512F and ACT-783R (Carbone & Kohn 1999) were used to amplify part of the actin gene (actA). The 28S large subunit nrDNA (LSU) was amplified using primers LR0R (Moncalvo et al. 1995) and LR5 (Vilgalys & Hester 1990). The RNA polymerase II second largest subunit (rpb2) was amplified with RPB2-5F2 (Sung et al. 2007) and fRPB2-7cR (Liu et al. 1999). Glyceraldehyde-3-phosphate dehydrogenase (gapdh) was amplified using primers Gpd1-LM and Gpd2-LM (Myllys et al. 2002). For P. citricarpa isolates the alternative primers Gpd1 (Guerber et al. 2003) and GPDHR2 (Glienke et al. 2011) were used to amplify gapdh. The PCR amplification mixtures and cycling conditions for ITS, actA, tef1, LSU and gapdh were followed as described by Glienke et al. (2011). Due to the lack of available rpb2 gene sequences of Phyllosticta isolates, we generated these sequences for all the strains used for this study (except for P. citrimaxima CPC 20276 = CBS 136059, culture has been lost). The rpb2 PCR was performed in a total volume of 25 μL and the mixture consisted of 1 μL genomic DNA, 1× PCR Buffer (Bioline GmbH, Luckenwalde, Germany), 0.75 μM MgCl2, 1.85 μM of each dNTP, 0.45 μM of each primer and 0.5 μL BioTaq Taq DNA polymerase (Bioline GmbH, Luckenwalde, Germany). A touchdown PCR protocol was used for rpb2: initial denaturation (94 °C, 5 min), five amplification cycles (94 °C, 45 s; 60 °C, 45 s; 72 °C, 2 min), five amplification cycles (94 °C, 45 s; 58 °C, 45 s; 72 °C, 2 min), 30 amplification cycles (94 °C, 45 s; 54 °C, 45 s; 72 °C, 2 min) and a final extension step (72 °C, 8 min). The PCR products were sequenced in both directions using the BigDye® Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems Life Technologies, Carlsbad, CA, USA), after which amplicons were purified through Sephadex G-50 Fine columns (GE Healthcare, Freiburg, Germany) in MultiScreen HV plates (Millipore, Billerica, MA). Purified sequence reactions were analysed on an Applied Biosystems 3730xl DNA Analyzer (Life Technologies, Carlsbad, CA, USA). The DNA sequences generated were analysed and consensus sequences were computed using the program SeqMan Pro (DNASTAR, Madison, WI, USA).
Phylogenetic analyses
Novel sequences generated in this study were queried against the NCBI's GenBank nucleotide database to determine the closest relatives for a taxonomic framework of the studied isolates. Alignments of different gene regions, including sequences obtained from this study and sequences downloaded from GenBank, were initially performed by using the MAFFT v. 7 online server (http://mafft.cbrc.jp/alignment/server/index.html) (Katoh & Standley 2013), and then manually adjusted in MEGA v. 6.06 (Tamura et al. 2013). Additional reference sequences were selected based on recent studies on Phyllosticta species (Glienke et al., 2011, Wang et al., 2012, Wikee et al., 2013a).
Phylogenetic analyses were based on both Bayesian Inference (BI) and Maximum Parsimony (MP) analyses. For BI, the best evolutionary model for each partition was determined using MrModeltest v. 2.3 (Nylander 2004) and incorporated into the analysis. MrBayes v. 3.2.5 (Ronquist et al. 2012) was used to generate phylogenetic trees under optimal criteria per partition. The Markov Chain Monte Carlo (MCMC) analysis used four chains and started from a random tree topology. The heating parameter was set to 0.2 and trees were sampled every 100 generations. Analyses stopped once the average standard deviation of split frequencies was below 0.01. The MP analysis was done using PAUP (Phylogenetic Analysis Using Parsimony, v. 4.0b10; Swofford 2003). Phylogenetic relationships were estimated by heuristic searches with 100 random addition sequences. Tree bisection-reconnection was used, with the branch swapping option set on “best trees” only with all characters weighted equally and alignment gaps treated as fifth state. Tree length (TL), consistency index (CI), retention index (RI) and rescaled consistence index (RC) were calculated for parsimony and bootstrap analysis (Hillis & Bull 1993) was based on 1 000 replications. Sequences generated in this study were deposited in GenBank (Table 2) and alignments and phylogenetic trees in TreeBASE (www.treebase.org). Nomenclatural novelties were deposited in MycoBank (Crous et al. 2004).
Taxonomy
A subset of isolates of the four Phyllosticta species collected in this study was morphologically characterised. After 14 d of incubation in the dark at 27 °C, the morphological characteristics were examined by mounting fungal structures in clear lactic acid and 30 measurements at ×1 000 magnification were determined for each isolate using a Zeiss Axioscope 2 microscope with interference contrast (DIC) optics. Colony colour and growth rate were established on MEA, potato dextrose agar (PDA) and OA according to Crous et al. (2009). Sporulation was induced on pine needle agar (PNA) (Smith et al. 1996) and synthetic nutrient-poor agar (SNA) under near UV-light. Colony colour was determined on MEA, OA and PDA using the colour charts of Rayner (1970). Colony growth rates were assessed on MEA, OA and PDA in 90 mm Petri plates at 9–39 °C at 3 °C intervals. Three plates were used for each culture/media and two measurements of colony diameter perpendicular to each were made after 3, 6, 9 and 12 d of incubation in the dark, after which averages were computed. For each species × growth medium × incubation time combination, data were normalised to the maximum growth observed for that combination. The combined dataset with relative growth values (0 = no growth, 1 = maximum growth) was subjected to non-linear regression using the BETE function: Y = (a × ((X − Tmin)/(Tmax − Tminx)) ˆ b × (1−((X − Tmin)/(Tmax − Tminx)) ˆ c (Analytis, 1977, Leggieri et al., 2017). Goodness of fit was determined through linear regression of the predicted against actual relative growth values.
Mating type identification
The mating types of P. citricarpa strains were determined based on PCR amplification of a diagnostic region from each mating type idiomorph by using four primers, MAT111F3 (5′-GCAATGTGGCAGCGCAATCC-3′) and MAT111R3 (5′-TCTGGACCATCGGACTCATC-3′) for MAT1-1-1, and MAT121F6 (5′-GATCGTGGCAGGAGGCTTTG-3′) and MAT121R6 (5′-AACGACCAGCGATCGGTAAG-3′) for MAT1-2-1 (Amorim et al. 2017). The same reaction mixtures were used for the amplification of both primers sets. A total volume of 12.5 μL containing 1 μL genomic DNA, 1× PCR Buffer (Bioline GmbH, Luckenwalde, Germany), 0.63 μM MgCl2, 0.7 μM of each dNTP, 0.25 μM of each primer and 0.5 μL BioTaq Taq DNA polymerase (Bioline GmbH, Luckenwalde, Germany), was used.
The PCR programme for the primers MAT111F3–MAT111R3 consisted of initial denaturation (94 °C, 3 min), 25 amplification cycles (94 °C, 30 s; 60 °C, 30 s; 72 °C, 1 min), and a final extension step (72 °C, 10 min). For the primers MAT121F6–MAT121R6, 30 amplification cycles (94 °C, 30 s; 55 °C, 30 s; 72 °C, 1 min) were used. The amplified fragments were separated by electrophoresis at 100 V for 25 min on a 1 % (w/v) agarose gel stained with GelRed™ (Biotium, Hayward, CA, USA), and viewed under ultra-violet light. Sizes of amplicons were determined against a HyperLadder™ I molecular marker (Bioline).
Genotyping of P. citricarpa isolates
Fifteen published polymorphic SSR markers (Wang et al., 2016, Carstens et al., 2017) were used to compare the genotypes of the P. citricarpa isolates found in this study with populations from Australia, Brazil, China, South Africa and the USA (Carstens et al. 2017). The primer labelling as well as the PCR reactions and cycling conditions were as previously described in Carstens et al. (2017). The SSR alleles were scored using Genemapper software v. 4 (Life Technologies). To determine the within-population genetic diversity the following were calculated in GenAlEx v. 6.5 (Peakall & Smouse, 2012): number of alleles (Na), number of effective alleles, number of private alleles, number of polymorphic loci and Nei's measure of gene diversity (Nei 1973). A zero value for Nei's gene diversity is an indication that there is no genetic diversity within the population. Isolates with identical alleles across all the loci were considered clones or multilocus genotypes (MLGs). For the allele-based genetic analyses, a per population clone-corrected dataset was used. To assess the genetic variation between the European populations and those from other continents, an analysis of molecular variance (AMOVA) was conducted. The statistical significance was tested using 999 permutations. In order to perform this analysis, the 12 P. citricarpa populations from Carstens et al. (2017) were included in the dataset. The AMOVA was performed in GenAlEx v. 6.5 (Peakall & Smouse 2012).
Pathogenicity
Two isolates of each of the four Phyllosticta species isolated from specimens collected in Europe (P. capitalensis: CPC 27825, CPC 27917; P. paracapitalensis: CPC 26517, CPC 26700; P. citricarpa: CPC 27909, CPC 27913; P. paracitricarpa: CPC 27169, CPC 27170), were inoculated into mature, untreated fruits of sweet orange (Citrus sinensis Osbeck), cultivar ‘Valencia’ (from Spain), following the method described by Perryman et al. (2014) to obtain indicative results about pathogenicity. Three fruits per replicate for each isolate were inoculated and were arranged in a randomised complete block design. Fruits were washed and surface disinfected by immersion for 10 min in 70 % ethanol, and rinsed twice in autoclaved water. A suspension of conidia (1.0 × 105 conidia/mL) was obtained from cultures grown on PDA for 15 d at 27 °C, and was injected, 100 mL at a time, into 12 inoculation points on the surface of oranges. The suspension was inoculated by inserting a hypodermic sterile needle into the albedo (the white pith area just below the peel), approx. 2 mm deep. Control fruits were inoculated with sterile water. The inoculation points on each fruit were labelled with a dot made with a permanent marker. The inoculated oranges were incubated in sterile plastic boxes at 20 °C, with 100 % relative humidity, under a lighting rig providing a 12 h photoperiod. Lesion development was evaluated 5, 10 and 25 d after inoculation. The inoculated fungi were re-isolated from any tissue showing lesions and the identity of the re-isolated fungi was confirmed by sequencing loci tef1 and LSU.
Results
Sampling and isolation
A total of 64 monosporic isolates resembling those of the genus Phyllosticta were collected. The Phyllosticta isolates were recovered from five species of Citrus at 11 different sites. Among them, 32 isolates were obtained from fresh leaves, 28 were associated with leaf litter and four with leaf spot symptoms (Table 2). During the surveys performed no CBS symptoms were observed.
Phylogenetic analyses
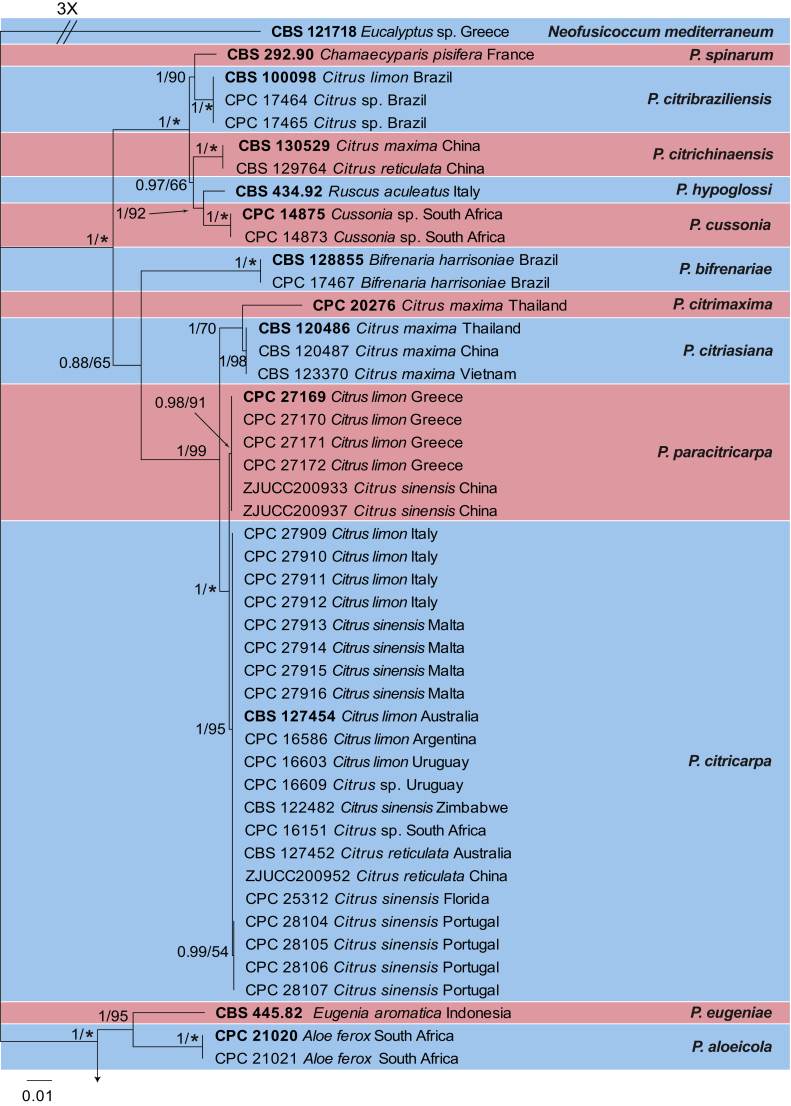
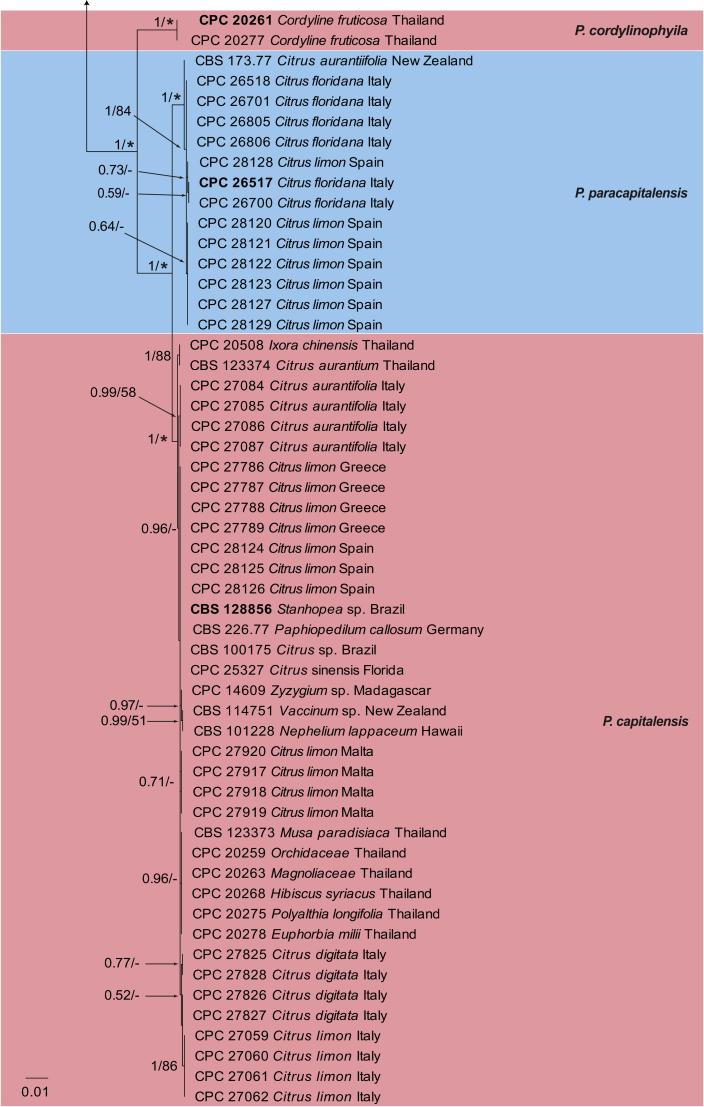
The combined species phylogeny of Phyllosticta consisted of 100 sequences, including the outgroup sequences of Neofusicoccum mediterraneum (culture CBS 121718). A total of 3 142 characters were included in the phylogenetic analyses; 693 characters were parsimony-informative, 315 were variable and parsimony-uninformative and 2 134 characters were constant. The maximum of 1 000 equally most parsimonious trees were saved (Tree length = 1 829, CI = 0.750, RI = 0.972 and RC = 0.729). Bootstrap support values from the parsimony analysis were plotted on the Bayesian phylogeny presented in Fig. 1. For the Bayesian analysis, MrModeltest suggested that the ITS partition should be analysed with a fixed state frequency distribution and all other loci with Dirichlet state frequency distributions. The following models were used in the Bayesian analysis: SYM+I+G (ITS), HKY+I (actA), GTR+G (tef1, gapdh, rpb2) and GTR+I (LSU).
Fig. 1.
Consensus phylogram resulting from a Bayesian analysis of the combined ITS, actA, tef1, gapdh, LSU and rpb2 sequence alignments. Bootstrap support values and Bayesian posterior probability values are indicated at the nodes. Substrate and country of origin, where known, are indicated next to the strain numbers. The tree was rooted to Neofusicoccum mediterraneum (CBS 121718).
In the Bayesian analysis, the ITS partition had 189 unique site patterns, the actA partition had 116 unique site patterns, the tef1 partition had 158 unique site patterns, the gapdh partition had 105 unique site patterns, the LSU partition had 76 unique site patterns, the rpb2 partition had 245 unique site patterns and the analysis ran for 1 900 000 generations, resulting in 38 002 trees of which 28 502 trees were used to calculate the posterior probabilities (Fig. 1). The main difference between the Bayesian and MP trees was the position of P. bifrenariae; in the Bayesian tree this species clustered basal to P. citricarpa whereas it was basal to the broader lineage containing the species clades of P. citricarpa to P. citribraziliensis in the parsimony analysis (data not shown). All other species clades were identical between the two analyses. The tree resolved 15 Phyllosticta species, two of which (P. paracapitalensis and P. paracitricarpa) are described as new in the Results – Taxonomy section.
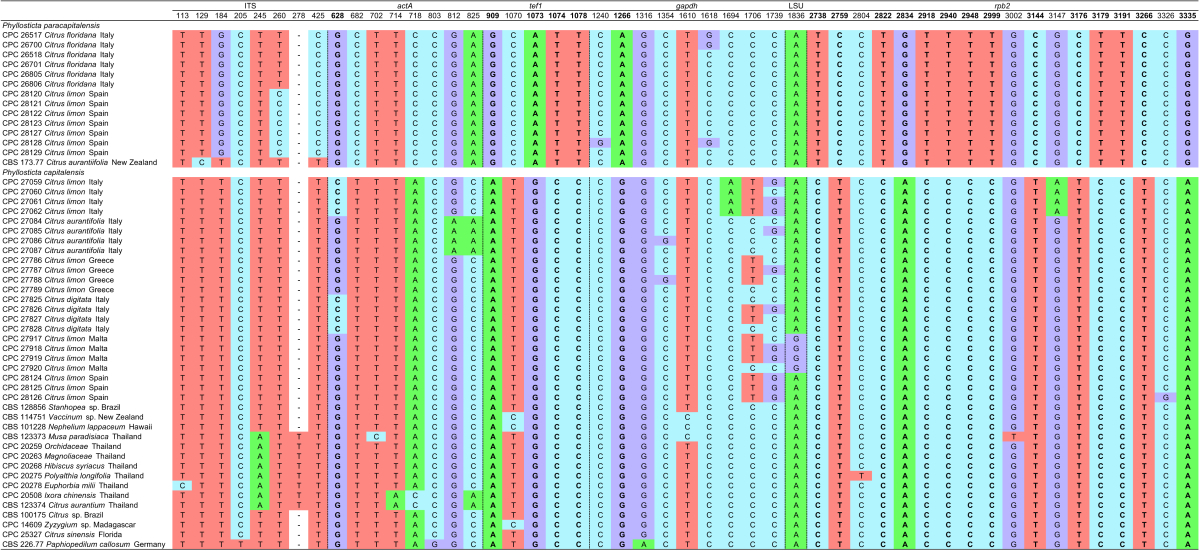
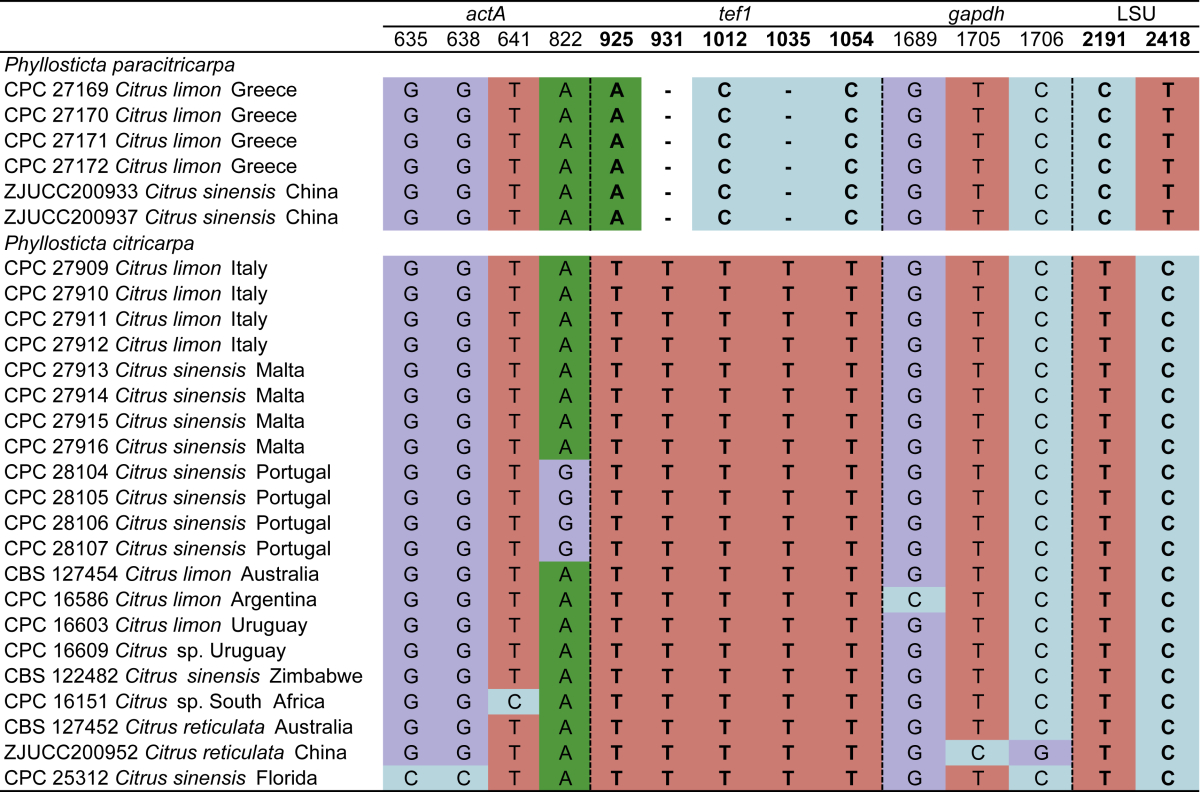
Nucleotide variations were observed in 49 base positions within the alignment of P. capitalensis isolates and those of the new species, P. paracapitalensis, included in this study (Table 3), and in 14 positions for P. citricarpa and the new species P. paracitricarpa (Table 4). Between the P. capitalensis and P. paracapitalensis clades, differences were present in all regions sequenced except for ITS. Specifically, 20 fixed nucleotide changes were observed over 3 142 nucleotides (one for actA, four for tef1, one for gapdh and 14 for rpb2). Moreover, seven fixed nucleotide changes were observed between P. citricarpa and P. paracitricarpa clades (five for tef1 and two for LSU). ITS, LSU and tef1 were sequenced to identify a further eight isolates of P. citricarpa (CPC 31171, CPC 31172, CPC 31173, CPC 31174, from Malta and CPC 31179, CPC 31180, CPC 31181, CPC 31182 from Portugal) and four isolates of P. paracitricarpa (CPC 31246, CPC 31247, CPC 31248, CPC 31249 from Greece) (data not shown).
Table 3.
Nucleotide differences observed among P. paracapitalensis and P. capitalensis isolates used in this study. Base positions include spaces caused by alignment gaps and refer to the position in the alignment deposited in TreeBASE. Base positions representing fixed nucleotide differences between the two species are in bold.
Table 4.
Nucleotide differences observed among P. paracitricarpa and P. citricarpa isolates used in this study. Base positions include spaces caused by alignment gaps and refer to the position in the alignment deposited in TreeBASE. Base positions representing fixed nucleotide differences between the two species are in bold.
Taxonomy
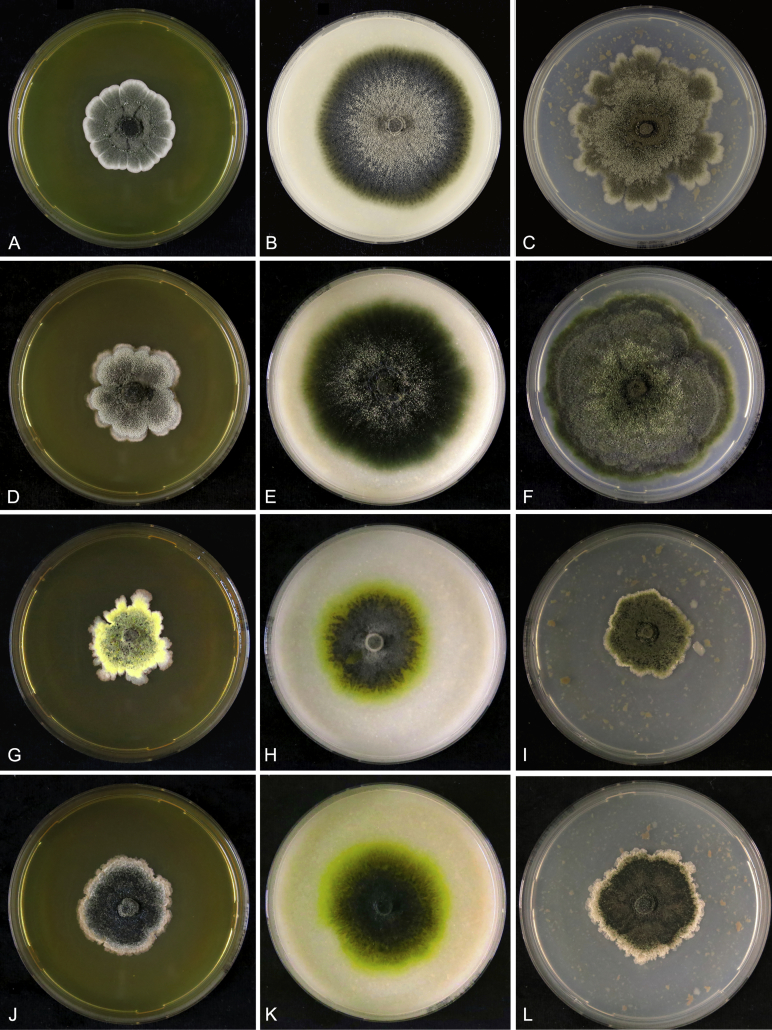
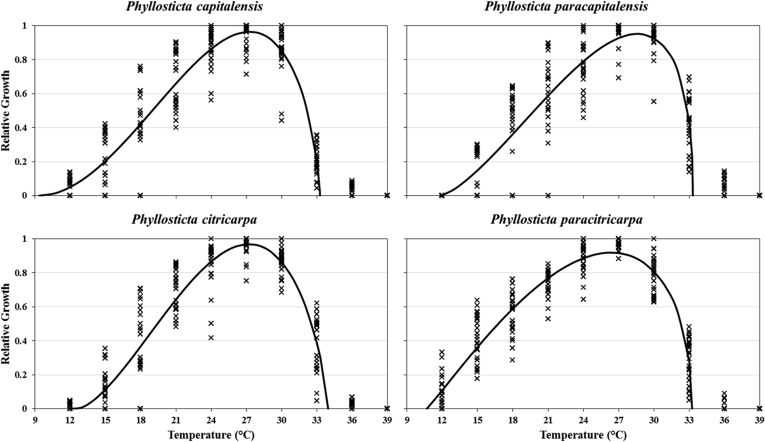
Morphological observations, supported by phylogenetic inference, were used to distinguish two known species (P. capitalensis and P. citricarpa) from two novel species. Culture characteristics were noted as dissimilar. The colour of upper and lower surfaces of Petri dishes were determined (Fig. 2). The BETE function fitted the relative growth data very well (R2 values 0.81 to 0.87) and predicted cardinal and optimal temperatures of 12.5–27.2–34.0 °C for P. citricarpa, 10.7–26.4–33.2 °C for P. paracitricarpa, 9.4–27.3–33.3 °C for P. capitalensis, and 11.8–28.6–33.3 °C for P. paracapitalensis (Fig. 3). After 9 d of incubation at 27 °C, P. capitalensis and P. paracapitalensis grew significantly faster (8.6–8.7 mm/d) on PDA and OA than P. citricarpa (4.8 and 6.6 mm/d, respectively) and P. paracitricarpa (4.0 and 5.4 mm/d, respectively), while growth of these species were significantly slower on MEA (5.7, 4.4, 4.5 and 3.3 mm/d, respectively). The isolates also differed morphologically from the other Phyllosticta species associated with citrus worldwide (Table 5). Based on the results of both the phylogenetic and morphological analyses, the two new species are described below.
Fig. 2.
Cultural characteristics of Phyllosticta species collected from citrus in Europe after 7 d at 27 °C on MEA, OA and PDA (respectively in 1st, 2nd and 3rd column). A–C.P. paracapitalensis. D–F.P. capitalensis. G–I.P. paracitricarpa. J–L.P. citricarpa.
Fig. 3.
Relative growth (0 to 1 scale) values on MEA, OA and PDA of Phyllosticta species collected in this study as influenced by incubation temperatures of 9–39 °C as fitted to a BETE function [Y = (a × ((X − Tmin)/(Tmax − Tminx)) ˆ b × (1−((X − Tmin)/(Tmax − Tminx)) ˆ c] with parameter values of a, Tmin, Tmax, b, c, and goodness of fit for P. capitalensis (8.942, 9.357, 33.261, 2.988, 0.665, R2 = 0.835), P. paracapitalensis (9.715, 11.820, 33.310, 3.551, 0.408, R2 = 0.806), P. citricarpa (6.932, 12.541, 33.962, 2.179, 0.749, R2 = 0.866) and P. paracitricarpa (6.281, 10.687, 33.247, 2.283, 0.471, R2 = 0.873).
Table 5.
Morphological characteristics of Phyllosticta spp. associated with citrus.
| Species | Ascomata |
Asci |
Ascospores |
Conidiomata |
Conidiogenous cells |
Conidia |
Spermatia |
Reference | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size (μm) | Shape | Size (μm) | Shape | Size (μm) | Shape | Size (μm) | Shape | Size (μm) | Shape | Size (μm) | Shape | Size (μm) | Shape | ||
| P. capitalensis | 250 | globose to pyriform | 58–80 × 11–15 | clavate | 15–17 × 5–6 | limoniform | 300 × 250 | globose to ampulliform | 7–10 × 3–5 | subcylindrical to ampulliform to doliiform | (10–)11–12(–14) × (5–)6–7 | ellipsoid to obovoid | – | – | Hennings (1908) |
| P. citriasiana | – | – | – | – | – | – | 120–240 × 125–225 | globose, subglobose to ellipsoidal | 7–17 × 3–5 | subcylindrical to ampulliform or doliiform | (10–)12–14(–16) × (5–)6–7(–8) | ellipsoid to obovoid | 3–5 × 1–2 | bacilliform to ellipsoid | Wulandari et al. (2009) |
| P. citribraziliensis | – | – | – | – | – | – | 250 | globose | 7–20 × 3–4 | subcylindrical to doliiform | 10–12 × 6–7 | ellipsoid to obovoid | – | – | Glienke et al. (2011) |
| P. citricarpa | – | – | – | – | – | – | 250 | globose to ampulliform | 7–12 × 3–4 | subcylindrical to doliiform | (10–)11–12(–14) × ( –)7(–8) | ellipsoid to obovoid | – | – | Van der Aa (1973) |
| P. citrichinaensis | 100–300 × 100–200 | subglobose to pyriform | 42–81 × 10–14 | subclavate to cylindrical | 14–20 × 7–8 | fusiform to ellipsoidal | 100–200 × 100–200 | globose or subglobose | 6–12 × 2–5 | lageniform | (7–)8–12(−13) × 6–9 | ellipsoid to obovoid | 7–9 × 1–2 | bacilliform | Wang et al. (2012) |
| P. citrimaxima | – | – | – | – | – | – | 150–160 × 120–130 | globose | 3–5 × 1–2 | cylindrical | 5(–8) × (3–)4(–7) | ellipsoid | – | – | Wikee et al., 2013a, Wikee et al., 2013b |
| P. paracapitalensis | up to 300 | globose | 40–75 × 10–12 | subcylindrical to clavate | 16–17 × 6 (–7) | limoniform | up to 250 | globose | 7–15 × 3–4 | subcylindrical | (9–)12–13(–14) × (6–)7 | ellipsoid to obovoid | – | – | This study |
| P. paracitricarpa | – | – | – | – | – | – | 250 | globose | 12–17 × 3–4 | subcylindrical | (9–)11–13(–15) × 7–8(–9) | ellipsoid to obovoid | – | – | This study |
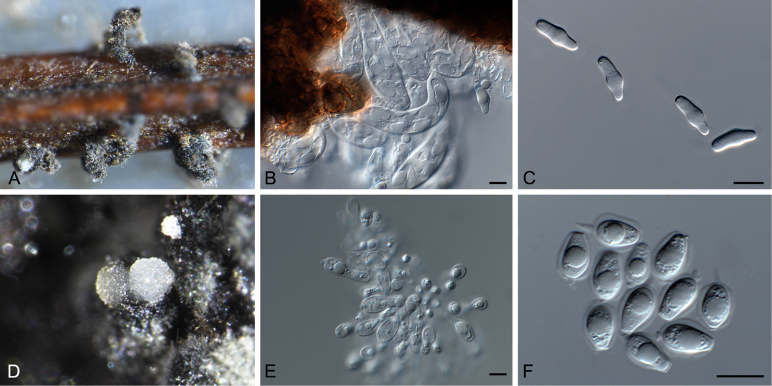
Phyllosticta paracapitalensis Guarnaccia & Crous, sp. nov. MycoBank MB817204; Fig. 4.
Fig. 4.
Phyllosticta paracapitalensis (CBS 141353). A. Ascomata forming on PNA. B. Asci with ascospores. C. Ascospores. D. Conidiomata forming on SNA. E. Conidiogenous cells giving rise to conidia. F. Conidia with mucoid sheaths and apical appendages. Scale bars = 10 μm.
Etymology: Named after its close morphological resemblance and phylogenetic relationship to P. capitalensis.
Conidiomata (on PNA) pycnidial, solitary, black, erumpent, globose, exuding colourless conidial masses; pycnidia up to 250 μm diam, elongated in culture on PNA; pycnidial wall of several layers of textura angularis, to 30 μm thick; inner wall of hyaline textura angularis. Ostiole central, to 20 μm diam. Conidiophores subcylindrical to ampulliform, reduced to conidiogenous cells, or with 1–2 supporting cell, that can be branched at the base, 7–20 × 4–6 μm. Conidiogenous cells terminal, subcylindrical, hyaline, smooth, coated in a mucoid layer, 7–15 × 3–4 μm; proliferating several times percurrently near apex. Conidia (9–)12–13(–14) × (6–)7 μm, solitary, hyaline, aseptate, thin and smooth-walled, granular, or with a single large central guttule, fusoid-ellipsoid, tapering towards a narrow truncate base, 3–4 μm diam, enclosed in a persistent mucoid sheath, 2–3 μm thick, and bearing a hyaline, apical mucoid appendage, (4–)5–7(–8) × 1.5(–2) μm, flexible, unbranched, tapering towards an acutely rounded tip. Ascomata solitary or in clusters of 2–3, erumpent, globose, up to 300 μm diam, with elongated neck to 500 μm long, with central ostiole; wall of 3–6 layers of brown textura angularis. Asci bitunicate, 8-spored, stipitate, with small pedicel and well developed apical chamber, hyaline, subcylindrical to clavate, 40–75 × 10–12 μm. Ascospores bi- to multiseriate, hyaline, smooth, granular with large central guttule, aseptate, straight, rarely curved, widest in the middle, limoniform with mucoid caps at obtuse ends, (15–)16–17(–18) × 6(–7) μm.
Culture characteristics: On MEA, colonies appear woolly, flat, irregular, initially white with abundant mycelium, gradually becoming greenish to dark green after 2–3 d with white hyphae on the undulate margin; reverse dark green to black. On OA, colonies appear flat with a regular margin, initially hyaline with abundant mycelium, gradually becoming dark greenish after 3–4 d; reverse dark green to black. On PDA, colonies appear irregular, woolly, initially white, gradually becoming greenish to dark green after 2–3 d with white hyphae on the undulate margin; reverse black. After 12 d in the dark at 27 °C, mycelium reached the edge of the Petri dish. The optimum growth rate was observed at 27 °C. No growth was observed at 12 °C and 39 °C.
Specimen examined: Italy, Sicily, on living leaf of Citrus × floridana, 4 Mar. 2015, V. Guarnaccia (holotype CBS H-22663, culture ex-type CPC 26517 = CBS 141353).
Notes: Phyllosticta paracapitalensis was isolated from leaves of Citrus limon and C. ×floridana as an endophyte. This species is similar to P. capitalensis, its sister species, but represents a distinct taxon, supported by molecular and morphological differences. Phyllosticta paracapitalensis differs from P. capitalensis in having longer ascomatal necks, narrower asci, and slightly larger ascospores. The asexual morph presents solitary and globose conidiomata that differ from those of P. capitalensis (aggregated and globose to ampuliform). Furthermore, the ostioles are larger and the conidiogenous cells are longer than P. paracapitalensis.
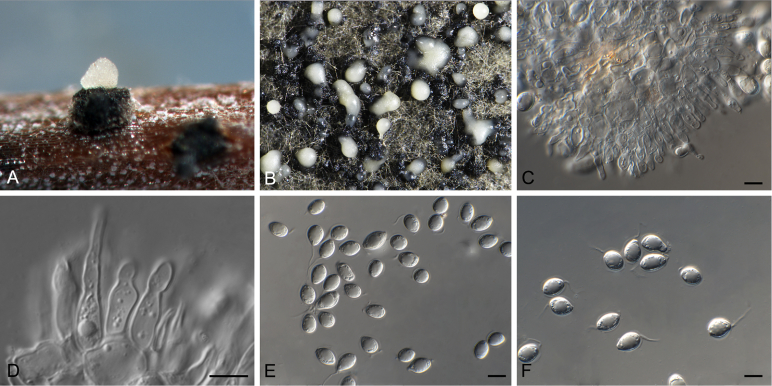
Phyllosticta paracitricarpa Guarnaccia & Crous, sp. nov. MycoBank MB817205. Fig. 5.
Fig. 5.
Phyllosticta paracitricarpa (CBS 141357). A, B. Conidiomata forming on PNA. C, D. Conidiogenous cells giving rise to conidia. E, F. Conidia with mucoid sheaths and apical appendages. Scale bars = 10 μm.
Etymology: Named after its close morphological resemblance and phylogenetic relationship to P. citricarpa.
Conidiomata (on PNA) pycnidial, solitary, black, erumpent, globose, exuding colourless conidial masses; pycnidia up to 250 μm diam, elongated in culture on PNA; pycnidial wall of several layers of textura angularis, 20–30 μm thick; inner wall of hyaline textura angularis. Ostiole central, up to 10 μm diam. Conidiophores subcylindrical to ampulliform, reduced to conidiogenous cells, or with 1–2 supporting cell, that can be branched at the base, 15–25 × 4–5 μm. Conidiogenous cells terminal, subcylindrical, hyaline, smooth, coated in a mucoid layer, 12–17 × 3–4 μm; proliferating several times percurrently near apex. Conidia (9–)11–13(–15) × 7–8(–9) μm, solitary, hyaline, aseptate, thin and smooth-walled, granular, or with a single large central guttule, ellipsoid to obovoid, tapering towards a narrow truncate base, 3–4 μm diam, enclosed in a thin persistent mucoid sheath, 1–1.5 μm thick, and bearing a hyaline, apical mucoid appendage, (8–)10–12(–15) × 1.5(–2) μm, flexible, unbranched, tapering towards an acutely rounded tip.
Culture characteristics: Colonies on MEA flat, with irregular edge; surface initially yellow becoming leaden grey in the centre, yellow at margin, and leaden grey underneath. On PDA colonies were flat, rather regular and slow growing, initially white-grey mycelium, gradually becoming greenish to dark green, with white hyphae at the margin; reverse black. On OA flat, spreading, olivaceous grey, becoming pale dark grey towards the margin, with sparse to moderate aerial mycelium; surrounded by a diffuse yellow pigment in the agar medium. After 12 d in the dark the optimum growth was observed at 27 °C on MEA, OA and PDA (33, 53 and 41 mm). No growth was observed at 9 °C and 39 °C.
Specimen examined: Greece, Mastro, on leaf litter of Citrus limon, 6 May 2015, V. Guarnaccia (holotype CBS H-22664, culture ex-type CPC 27169 = CBS 141357).
Notes: Phyllosticta paracitricarpa was isolated from Citrus limon leaf litter in Europe (this study) and from lesions on C. sinensis fruits in China (Wang et al. 2012). This species is similar to P. citricarpa, its sister species, but represents a distinct taxon, based on phylogenetic analyses and morphological differences. Phyllosticta paracitricarpa differs from P. citricarpa in having longer and slightly narrower conidiophores, larger conidiogenous cells and conidia. Phyllosticta paracitricarpa colonies on MEA appear yellow becoming leaden-grey in the centre, and yellow at the margin, different from P. citricarpa colonies that are olivaceous grey.
Mating type identification of P. citricarpa
The Phyllosticta mating type primer sets were successful in amplifying the respective portions of the MAT1-1-1 or the MAT1-2-1 idiomorphs of the 21 P. citricarpa isolates tested (Table 2). The primer pair MAT111F3–MAT111R3 amplified a fragment of approximately 606 bp in eight isolates, and the primer pair MAT121F6–MAT121R6 amplified 692-bp-fragments in the remaining 13 isolates.
Genotyping of P. citricarpa isolates
The 20 P. citricarpa isolates from four localities in three countries (Malta, Italy and Portugal) were regarded as four “putative” populations (due to the low number of isolates obtained and the sampling strategy employed) and were genotyped with the 15 SSR markers. Among the 20 isolates that were analysed, only two MLGs were identified. The two populations from Malta and the population from Italy shared a single MLG; the other MLG was identified in the population from Portugal. None of the 15 SSR markers were polymorphic in the populations from Italy, Malta and Portugal and therefore indicated very low gene diversity in the populations (0.000; results not shown). The population from Portugal shared its single MLG with all three populations from Australia, while the populations from Italy and Malta shared one MLG, which was not shared with any of the populations from Australia, Brazil, China, Portugal, South Africa or the USA. For the AMOVA analyses, the data from the three populations from Italy and Malta were combined as one population (Italy+Malta) as these three populations shared one MLG. Pairwise PhiPT values (Table 6) indicated that the Portugal population was genetically significantly (P ≤ 0.05) differentiated from the China (PhiPT = 0.634; P = 0.001), Italy+Malta (PhiPT = 1.000; P = 0.001), South Africa (PhiPT = 0.311; P = 0.002), and the USA (PhiPT = 1.000; P = 0.001) populations. The Portugal population was not significantly differentiated from the Australia population (PhiPT = 0.000; P = 0.418), and also not from the Brazil population (PhiPT = 0.322; P = 0.155). The Italy+Malta population was significantly (P ≤ 0.05) differentiated from the Australia (PhiPT = 0.258; P = 0.001), China (PhiPT = 0.651; P = 0.002), South Africa (PhiPT = 0.365; P = 0.002), Brazil (PhiPT = 0.483; P = 0.043), the USA (PhiPT = 1.000; P = 0.001) and Portugal (PhiPT = 1.000; P = 0.001) populations.
Table 6.
Pairwise PhiPT values (below the diagonal) averaged over 15 microsatellite loci of Phyllosticta citricarpa populations from Australia, Brazil, China, Italy+Malta, Portugal, South Africa and the United States. Significance P-values are indicated above the diagonal.
| Australia | Brazil | China | Italy + Malta | Portugal | South Africa | USA | |
|---|---|---|---|---|---|---|---|
| Australia | – | 0.011 | 0.001 | 0.001 | 0.418 | 0.001 | 0.422 |
| Brazil | 0.097 | – | 0.001 | 0.043 | 0.155 | 0.313 | 0.342 |
| China | 0.649 | 0.659 | – | 0.002 | 0.001 | 0.001 | 0.001 |
| Italy + Malta | 0.258 | 0.483 | 0.651 | – | 0.001 | 0.002 | 0.001 |
| Portugal | 0.000 | 0.322 | 0.634 | 1.000 | – | 0.002 | 0.001 |
| South Africa | 0.165 | 0.013 | 0.700 | 0.365 | 0.311 | – | 0.452 |
| USA | 0.000 | 0.013 | 0.674 | 1.000 | 1.000 | 0.000 | – |
Pathogenicity
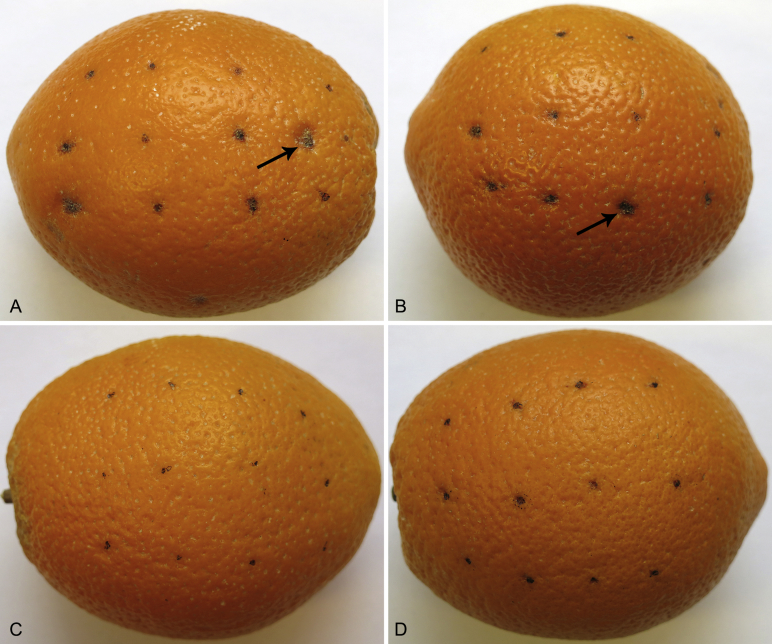
After 25 d, some inoculation points (approx. 75 %) showed atypical lesions. The lesions developed only on fruits inoculated with P. citricarpa (CPC 27909, CPC 27913) and P. paracitricarpa isolates (CPC 27169, CPC 27170). No lesions were observed on fruits inoculated with P. capitalensis (CPC 27825, CPC 27917), P. paracapitalensis (CPC 26517, CPC 26700) (Fig. 6), or on control fruits (not shown). The lesions caused by P. citricarpa and P. paracitricarpa were similar (Fig. 6). The latter species were consistently re-isolated from the fruit lesions, albeit from lesions atypical of the CBS disease, and identified by sequencing and comparing the loci tef1 and LSU.
Fig. 6.
Fruit of Citrus sinensis (‘Valencia’) artificially inoculated with Phyllosticta spp. A. Lesions caused by P. citricarpa. B. Lesions caused by P. paracitricarpa. C, D. No symptoms were observed on fruits inoculated with P. capitalensis and P. paracapitalensis.
Discussion
Phylogenetic studies published on the genus Phyllosticta in recent years have substantially reshaped its taxonomy (Glienke et al., 2011, Wang et al., 2012, Wikee et al., 2013a). The present study represents the first results of fresh collections of several Phyllosticta isolates and species associated with citrus in Europe, and the first DNA sequence analyses of strains from almost all continents.
Phyllosticta capitalensis has been recorded worldwide as a common endophyte of diverse host plants (Baayen et al. 2002). Phyllosticta citricarpa is confined to Citrus species on which it causes CBS in summer rainfall citrus growing areas in several countries. Despite the fact that these two species are morphologically distinct, their identification has often been confused (Everett & Rees-George 2006). Conidia of P. citricarpa (11–12 × 7 μm) are similar to those of P. capitalensis (11–12 × 6–7 μm), but have a thinner mucoid sheath. Moreover, P. citricarpa strains produce a distinct yellow pigment on OA, and are slower growing than P. capitalensis. The most recent studies focussing on the taxonomy of Phyllosticta species showed the occurrence of additional species associated with Citrus. Glienke et al. (2011) described P. citribraziliensis from healthy leaves. An additional three species were reported as Citrus pathogens in Asia: P. citriasiana and P. citrimaxima cause Citrus Tan Spot on pomelo fruits (Wulandari et al., 2009, Wikee et al., 2013a) and P. citrichinaensis causes a brown spot and red-brown protuberant freckle on citrus leaves and fruits (Wang et al. 2012).
Citrus Black Spot and symptoms similar to that caused by P. citricarpa, P. citriasiana, P. citrimaxima and P. citrichinaensis have never been reported in citrus-producing European countries (European Union, 1998, Kotzé, 2000). Climatic conditions play a primary role in the ability of P. citricarpa to establish and to cause CBS disease, most notably warm summer rainfall conditions that would allow spore production, dissemination and infection during periods of fruit susceptibility (Kiely, 1948a, Kiely, 1948b, Kotzé, 1963, Kotzé, 1981, McOnie, 1967, McOnie, 1964, Huang and Chang, 1972, Lee and Huang, 1973, Noronha, 2002, Fourie et al., 2013, Yonow et al., 2013, Magarey et al., 2015).
Given the long history of trade in citrus propagation material between Europe and Asia, where CBS is endemic and also regarded as the centre of origin of citrus, (Ramón-Laca, 2003, Mabberley, 2004, Nicolosi, 2007), and the potential for illegal movement of plant propagating material, the likely coincidental spread of citrus-specific Phyllosticta species to Europe could reasonably be expected. To investigate this possibility, several surveys were carried out during this study, resulting in the collection of 64 Phyllosticta isolates. A subset of 52 European isolates were compared to several reference isolates using partial gene sequences of six different loci, as well as morphological characteristics. Based on a comparison with sequences retrieved from GenBank of an additional 43 strains (Glienke et al., 2011, Wang et al., 2012, Wikee et al., 2013a), four distinct Phyllosticta species, including two new species, were delineated from several Citrus species growing in five European countries.
The distribution of the Phyllosticta species isolated in this study varied in terms of host and tissue type from which they were recovered. Phyllosticta capitalensis was recovered in all countries sampled and P. paracapitalensis in Italy and Spain only. Both species were isolated from asymptomatic leaves. Phyllosticta citricarpa and P. paracitricarpa were isolated from leaf litter only. Phyllosticta citricarpa was found in Italy, Malta and Portugal, whereas P. paracitricarpa was isolated only from samples collected in Greece. Phyllosticta capitalensis was associated with P. paracapitalensis in the same specimens collected in Spain, but in this survey P. citricarpa and P. paracitricarpa were not found associated with P. capitalensis.
Wang et al. (2012) reported two sub-clades (I and II) of P. citricarpa associated with Citrus spp. in China by comparison of ITS, actA and tef1 sequences data. In this study, we used partial regions of an additional three loci, and fixed nucleotide differences were observed within the tef1 and LSU regions, supporting the splitting of the “P. citricarpa” clade in two taxa: P. citricarpa s.str. and the new species P. paracitricarpa. Moreover, this study establishes the presence of P. paracitricarpa only in Asia and Europe and represents the first report of P. citricarpa in Europe. Phyllosticta paracitricarpa was isolated from fruit lesions in China and caused lesions on citrus fruit in the pathogenicity test performed in this study. Further surveys and research is required to determine the importance of P. paracitricarpa as a citrus pathogen.
The origin of P. citricarpa in Europe is not clear at present. On a genotypic level, the P. citricarpa populations from Italy+Malta and Portugal represented two respective clones, differing from each other in both their MLGs and mating types. These populations further differed from one another in their degree of connectivity and differentiation from the other populations from Australia, Brazil, China, South Africa and the USA. Analysis of molecular variance showed that populations from Portugal and Australia are more strongly connected to each other than to other populations. Interestingly, “Lisbon” lemon was introduced into Australia from Portugal in 1824 (Morton 1987), while CBS was first described in Australia in 1895 (Benson 1895). Very little connectivity was evident between the Portuguese population and those from the other continents, including the population from Italy+Malta. Also, the Italy+Malta population seemed to be distinct from the other populations. These findings suggest two separate introductions into Europe. However, in order to determine whether there were other introductions of P. citricarpa into Europe and to infer the origin of these introductions, additional populations from Europe, Asia and the Oceania countries need to be studied. The description of P. paracitricarpa from Greece and China suggests connectivity in this species with Asia.
No evidence of CBS disease in European citrus trees was observed in this study. The P. citricarpa isolates were found in leaf litter of old C. limon and C. sinensis trees (20 to 60 years old) that were situated in gardens, and not found in any of the commercial orchards or nurseries surveyed. Fruit is not considered a pathway for spread (USDA APHIS 2010) and evidence that might suggest a fruit pathway (such as nearby compost heap, waste disposal or processing plants; Baker et al. 2014) was not observed. Movement of infected plant material is regarded as the most likely means of long-distance spread of P. citricarpa (Kiely, 1948b, Kotzé, 1981). Whilst import of citrus plants for planting is presently not permitted, unless it is plant propagation material that is handled through appropriate quarantine procedures, the introduction of P. citricarpa found in Portugal, Malta and Italy therefore most likely occurred via the introduction of plants many years ago or via illegal movement of such plants.
Phyllosticta citricarpa was found at very low frequency only in a few of the sites investigated, while P. paracitricarpa was found only at one site in Greece. CBS disease symptoms were never observed. Our results indicate that the presence of P. citricarpa and P. paracitricarpa is not associated with disease under European climatic conditions.
Twenty-three P. capitalensis strains were isolated as endophyte from leaves of four Citrus species collected. This taxon can occur in fruit or leaf lesions caused by other fungi or insects (Wikee et al. 2013b). Indeed, in this study, P. capitalensis was found associated with leaf lesions (caused by insects) of the ornamental C. medica var. sarcodactylis. Wikee et al. (2013a) indicated that the phylogeny of Phyllosticta derived from the ITS and actA genomic loci is sufficiently robust to differentiate most taxa, except those closely related to P. capitalensis. In our study, sequences of a partial region of rpb2 of Phyllosticta spp. helped to resolve differences in nucleotides within P. capitalensis. Moreover, fixed nucleotide differences were observed in tef1, demonstrating the separation of the new species P. paracapitalensis with highly supported independent lineages in the phylogenetic tree. Phyllosticta paracapitalensis was isolated as endophyte from commercial orchards of C. limon in Spain and from C. floridana cultivated in ornamental plant nurseries in Italy. One strain (CBS 173.77) isolated from C. aurantiifolia in New Zealand during February 1974, previously identified as P. capitalensis, grouped with the European isolates of P. paracapitalensis in the present phylogenetic analyses. Further studies must be conducted on a wider global selection of strains to clarify its host association and distribution.
Morphological characteristics of isolates grown on several media were consistent with those already reported in literature (Baayen et al., 2002, Glienke et al., 2011, Wikee et al., 2013a). Optimal temperatures for P. citricarpa (27.2 °C) and P. capitalensis (27.3 °C) predicted from the BETE function fitted to the relative growth data were similar to those reported by previous studies (Kotzé, 1981, Er et al., 2014), but cardinal temperatures were more contracted with Tmin of (12.5 and 9.4 °C, respectively). Optimal temperatures for P. paracitricarpa and P. paracapitalensis were lower (26.4 °C) and higher (28.6 °C), respectively. Growth rates of P. capitalensis and P. paracapitalensis were similar and significantly faster than those of P. citricarpa and P. paracitricarpa.
Results of this study showed that two (P. citricarpa and P. paracitricarpa) of the four species isolated from specimens collected in Europe induced atypical lesions (necrosis) in artificially inoculated mature sweet orange fruit and could be re-isolated from these lesions, while P. capitalensis and P. paracapitalensis induced no lesions. From this assay, it appears that P. paracapitalensis is similar to P. capitalensis, demonstrating them to have similar ecologies, occurring as asymptomatic endophytes in citrus tissue. Considering that mature citrus fruit are resistant to P. citricarpa infection under field conditions (Kiely, 1948b, Schutte et al., 2003, Schutte et al., 2012, Miles et al., 2004), and since the harsh artificial inoculation technique used in the pathogenicity assay did not resemble natural field infection (i.e. direct penetration of unwounded tissue following long wetness periods; Kotzé, 1963, McOnie, 1967, Noronha, 2002) these findings should be regarded as preliminary. Phyllosticta paracitricarpa caused similar lesions to those caused by P. citricarpa in this assay and appears to be pathogenic, which is supported by its isolation from lesions on fruit in China, but further surveys are required to elucidate.
Including the two taxa newly described in this study, eight Phyllosticta species are now associated with citrus: P. citricarpa and P. capitalensis are present on all continents where citrus is cultivated, P. paracapitalensis is reported in Europe and New Zealand, while P. paracitricarpa is present in Asia and Europe. As previously published by several authors, the pathogenic P. citrichinaensis, P. citriasiana and P. citrimaxima are present only in Asia, and the endophyte P. citribraziliensis has been isolated only in South America (Wulandari et al., 2009, Glienke et al., 2011, Wang et al., 2012, Wikee et al., 2013a). The presence in Europe of both P. citricarpa and P. paracitricarpa was not associated with any visible signs of infection; indeed, neither CBS or Citrus Tan Spot have ever been reported or observed during the extensive surveys performed in the present study.
Recent studies performed in Florida, USA (Zhang et al., 2015, Wang et al., 2016), supported the heterotallism of P. citricarpa, finding only MAT1-2-1 isolates present in Florida (based on 113 isolates) while 26 strains from Australia displayed an equal ratio of the two mating types. Amorim et al. (2017) recently showed that in Brazil the two idiomorphs occur in a 1:1 ratio. Furthermore, Tran et al. (2017) reported for the first time the successful mating in vitro of P. citricarpa, confirming that this species is heterothallic and requires isolates of different MAT idiomorphs to be in direct physical contact for mating and production of sexual fruiting bodies. We found both MAT1-1-1 and MAT1-2-1 isolates present in Europe, but both mating types were not recovered together in the same country, indicating separate introductions that have not spread and remained isolated. A broader sampling is required, however, to determine whether this holds up when a larger population per area is sampled.
This study contributed significantly towards our understanding of the genotypic variation in P. capitalensis and P. citricarpa, splitting both groups into different taxa, and clearly showing that a multi-locus approach works well for distinguishing these species. The use of a three-gene analysis (ITS, actA, tef1) performed in a previous study (Wang et al. 2012) showed two poorly supported subclades within P. citricarpa. We used a further three genomic loci (gapdh, LSU and rpb2) to confirm that the two subclades actually represent two distinct species.
In this study we established the presence of P. citricarpa and the similar new species, P. paracitricarpa, for the first time in Europe. In spite of the occurrence of these species, neither was associated with disease symptoms, evidently because of unfavourable climatic conditions (Yonow et al., 2013, Magarey et al., 2015). Whilst it appears that these fungi were introduced with plant material many years ago, they apparently persist under these unfavourable conditions, most likely endophytically, and possibly through asexual reproduction. The latter hypothesis is supported by the finding that only one mating type was found per locality, and that some P. citricarpa pycnidiospore infection events were predicted to occur in these regions (Magarey et al. 2015). The number of suitable infection periods was, however, markedly fewer than those for regions where P. citricarpa causes CBS disease. Magarey et al. (2015) doubted the ability of P. citricarpa to persist and cause disease at a location where there is a low frequency of suitable seasons. Likewise, the climate suitability modelling conducted by Paul et al. (2005) and Yonow et al. (2013), indicated climatic unsuitability across the EU, with the exception of a few isolated areas around the Mediterranean Sea, where marginally suitable climatic conditions can be found. All these climate modelling studies were calibrated for climate suitability according to the presence, absence, distribution and severity of CBS disease, and not the potential presence of the fungus in the absence of disease. The findings from our study indicate that P. citricarpa was able to persist but did not induce CBS symptoms or spread, considering that it was found in only a few of the sites investigated and at very low frequency.
Acknowledgements
The authors are grateful to Arien van Iperen (cultures), Marjan Vermaas (photo plates) and Mieke Starink-Willemse (DNA isolation, amplification, and sequencing) for their technical assistance, to Ariena van Bruggen, (University of Florida) for sharing some strains, and to Tian Schutte for sharing his experience in surveying for CBS disease symptoms.
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Contributor Information
V. Guarnaccia, Email: v.guarnaccia@westerdijkinstitute.nl.
P.W. Crous, Email: p.crous@westerdijkinstitute.nl.
References
- Aa HA van der. Studies in Phyllosticta I. Studies in Mycology. 1973;5:1–110. [Google Scholar]
- Aa HA van der, Vanev S. Westerdijk Fungal Biodiversity Institute; Utrecht, The Netherlands: 2002. A revision of the species described in Phyllosticta. [Google Scholar]
- Aiello D., Carrieri R., Guarnaccia V. Characterization and pathogenicity of Colletotrichum gloeosporioides and C. karstii causing preharvest disease on Citrus sinensis in Italy. Journal of Phytopathology. 2015;163:168–177. [Google Scholar]
- Amorim R., Savi D.C., Ferreira-Maba L. MAT gene idiomorphs suggest a heterothallic sexual cycle in the citrus pathogen Phyllosticta citricarpa. European Journal of Plant Pathology. 2017;147:325–337. [Google Scholar]
- Analytis S. Uber die relation zwischen biologischer entwicklung und temperatur bei phytopathogenen pilzen. Phytopathologische Zeitschrift. 1977;90:64–76. [Google Scholar]
- Baayen R., Bonants P., Verkley G.J.M. Nonpathogenic isolates of the citrus black spot fungus, Guignardia citricarpa, identified as a cosmopolitan endophyte of woody plants, G. mangiferae (Phyllosticta capitalensis) Phytopathology. 2002;92:464–477. doi: 10.1094/PHYTO.2002.92.5.464. [DOI] [PubMed] [Google Scholar]
- Baker R., Bragard C., Candresse T. Scientific Opinion on the risk of Phyllosticta citricarpa (Guignardia citricarpa) for the EU territory with identification and evaluation of risk reduction options. EFSA Journal. 2014;12:3557. [Google Scholar]
- Baldassari R.B., Wickert E., de Goes A. Pathogenicity, colony morphology and diversity of isolates of Guignardia citricarpa and G. mangiferae isolated from Citrus spp. European Journal of Plant Pathology. 2008;120:103–110. [Google Scholar]
- Barr M.E. Some amerosporous ascomycetes on Ericaceae and Empetraceae. Mycologia. 1970;62:377–394. [Google Scholar]
- Barr M.E. Preliminary studies on the Dothideales in temperate North America. Contributions from the University of Michigan Herbarium. 1972;9:523–638. [Google Scholar]
- Bellotte J.A.M., Kupper K.C., Rinaldo D. Acceleration of the decomposition of Sicilian lemon leaves as an auxiliary measure in the control of citrus black spot. Tropical Plant Pathology. 2009;34:71–76. [Google Scholar]
- Benson A.H. Black spot of the orange. The Agricultural Gazette of New South Wales. 1895;4:249–252. [Google Scholar]
- Bezerra J.P.D., Santos M.G.S., Svedese V.M. Richness of endophytic fungi isolated from Opuntia ficus-indica Mill. (Cactaceae) and preliminary screening for enzyme production. World Journal Microbiology Biotechnology. 2012;28:1989–1995. doi: 10.1007/s11274-011-1001-2. [DOI] [PubMed] [Google Scholar]
- Bissett J. Discochora yuccae sp. nov. with Phyllosticta and Leptodothiorella synanamorphs. Canadian Journal of Botany. 1986;64:1720–1726. [Google Scholar]
- Brentu F.C., Oduro K.A., Offei S.K. Crop loss, etiology, and epidemiology of citrus black spot in Ghana. European Journal of Plant Pathology. 2012;133:657–670. [Google Scholar]
- Broadbent P. Quarantine in relation to Australian citrus imports and exports. Australasian Plant Pathology. 1995;24:145–156. [Google Scholar]
- Brodrick H.T. Department of Microbiology and Plant Pathology, University of Pretoria; 1969. Physiological studies with Guignardia citricarpa Kiely. D.Sc. Thesis. [Google Scholar]
- Calavan E.C. Black spot of citrus. Citrus Grower. 1960;323:11–15. [Google Scholar]
- Carbone I., Kohn L.M. A method for designing primer sets for the speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. [Google Scholar]
- Carstens E., Linde C.C., Slabbert R. A global perspective on the population structure and reproductive system of Phyllosticta citricarpa. Phytopathology. 2017 doi: 10.1094/PHYTO-08-16-0292-R. [DOI] [PubMed] [Google Scholar]
- Crous P.W., Gams W., Stalpers J.A. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology. 2004;50:19–22. [Google Scholar]
- Crous P.W., Slippers B., Wingfield M.J. Phylogenetic lineages in the Botryosphaeriaceae. Studies in Mycology. 2006;55:235–253. doi: 10.3114/sim.55.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Summerell B.A., Shivas R.G. Fungal Planet description sheets: 107–127. Persoonia. 2012;28:138–182. doi: 10.3767/003158512X652633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Verkley G.J.M., Groenewald J.Z., editors. Fungal Biodiversity. CBS Laboratory Manual Series 1. Westerdijk Fungal Biodiversity Institute; Utrecht, The Netherlands: 2009. [Google Scholar]
- De Hoog G.S., Gerrits van den Ende A.H.G. Molecular diagnostics of clinical strains of filamentous basidiomycetes. Mycoses. 1998;41:183–189. doi: 10.1111/j.1439-0507.1998.tb00321.x. [DOI] [PubMed] [Google Scholar]
- Dewdney M.M., Schubert T.S., Estes M.R. University of Florida; 2012. Florida citrus pest management guide: Citrus Black Spot. IFAS Extension PP279. [Google Scholar]
- Doidge E.M. Some diseases of Citrus prevalent in South Africa. South African Journal of Science. 1929;26:324. [Google Scholar]
- Donk M.A. Report of the committee for Fungi and Lichen 1964–1968. Taxon. 1968;17:578–581. [Google Scholar]
- Dummel D.M., Agostini J.P., Moschini R. Predictive model for ascospore release of Guignardia citricarpa using climatological data. Proceedings of the XIIth International Citrus Congress, Valencia, Spain, B. Sabater-Munoz et al. Acta Horticulturae. 2015;1065:953–963. [Google Scholar]
- EC 2000/29/EC Council Directive on protective measures against the introduction into the Community of organisms harmful to plants and plant products and against their spread within the Community. Official Journal of the European Communities L. 2000;169:1–112. [Google Scholar]
- Er H.L., Hendricks K., Goss E.M. Isolation and biological characterization of Guignardia species from citrus in Florida. Journal of Plant Pathology. 2014;96:43–55. [Google Scholar]
- European Union Commission Decision of 8 January 1998 recognizing certain third countries and certain areas of third countries as being free of Xanthomonas campestris (all strains pathogenic to Citrus), Cercospora angolensis Carv. et Mendes and Guignardia citricarpa Kiely (all strains pathogenic to Citrus) Official Journal of the European Communities. 1998;15:41–42. [Google Scholar]
- European Union . 2000. Final report of a mission carried out in Brazil from 3 to 6 July 2000 in order to evaluate the pre-export inspections on citrus fruit originating in Brazil and exported to the European Union.http://ec.europa.eu/food/fs/inspections/pi/reports/brazil/pi_rep_braz_1180-2000_en.pdf [Google Scholar]
- Everett K.R., Rees-George J. Reclassification of an isolate of Guignardia citricarpa from New Zealand as Guignardia mangiferae by sequence analysis. Plant Pathology. 2006;55:194–199. [Google Scholar]
- Fourie P., Schutte T., Serfontein S. Modeling the effect of temperature and wetness on Guignardia pseudothecium maturation and ascospore release in citrus orchards. Phytopathology. 2013;103:281–292. doi: 10.1094/PHYTO-07-11-0194. [DOI] [PubMed] [Google Scholar]
- Garran S.M. Citrus Black spot in the North East of Entre Rios: etiology epidemiology and control. Proceedings of the International Society of Citriculture. 1996:466–471. [Google Scholar]
- Glienke C., Pereira O., Stringari D. Endophytic and pathogenic Phyllosticta species, with reference to those associated with Citrus Black Spot. Persoonia. 2011;26:47–56. doi: 10.3767/003158511X569169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glienke-Blanco C., Aguilar-Vildoso C.I., Vieira M.L.C. Genetic variability in the endophytic fungus Guignardia citricarpa isolated from citrus plants. Genetics and Molecular Biology. 2002;25:251–255. [Google Scholar]
- Guarnaccia V., Groenewald J.Z., Polizzi G. High species diversity in Colletotrichum associated with citrus diseases in Europe. Persoonia. 2017;39:32–50. doi: 10.3767/persoonia.2017.39.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerber J.C., Liu B., Correll J.C. Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia. 2003;95:872–895. [PubMed] [Google Scholar]
- Hawksworth D.L., Crous P.W., Redhead S.A. The Amsterdam declaration on fungal nomenclature. IMA Fungus. 2011;2:105–112. doi: 10.5598/imafungus.2011.02.01.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennings P. Fungi S. Paulenses IV a cl. Puttemans collecti. Hedwigia. 1908;48:1–20. [Google Scholar]
- Hillis D.M., Bull J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology. 1993;42:182–192. [Google Scholar]
- Hu J., Johnson E.G., Wang N.Y. qPCR quantification of pathogenic Guignardia citricarpa and nonpathogenic G. mangiferae in Citrus. Plant Disease. 2014;98:112–120. doi: 10.1094/PDIS-04-13-0465-RE. [DOI] [PubMed] [Google Scholar]
- Huang C.S., Chang S.L. Leaf infection with citrus black spot and perithecial development in relation to ascospore discharge of Guignardia citricarpa Kiely. Journal of Taiwan Agricultural Research. 1972;21:256–263. [Google Scholar]
- Huang W.Y., Cai Y.Z., Hyde K.D. Biodiversity of endophytic fungi associated with 29 traditional Chinese medicinal plants. Fungal Diversity. 2008;33:61–75. [Google Scholar]
- Johnston P.R. Leaf endophytes of manuka (Leptospermum scoparium) Mycological Research. 1998;102:1009–1016. [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiely T.B. Preliminary studies on Guignardia citricarpa (n. sp.), the ascigerous stage of Phoma citricarpa McAlp., and its relation to black spot of citrus. Proceedings of the Linnean Society of New South Wales. 1948;73:249–292. [Google Scholar]
- Kiely T.B. Guignardia citricarpa (n.sp.) and its relationship to the black spot disease of Citrus in coastal orchards of New South Wales. Journal of the Australian Institute of Agriculture Science. 1948;14:81–83. [Google Scholar]
- Kiely T.B. Black spot of citrus in New South Wales coastal orchards. The Agricultural Gazette of New South Wales. 1949;60:17–20. [Google Scholar]
- Kotzé J.M. University of Pretoria; South Africa: 1963. Studies on the black spot disease of citrus caused by Guignardia citricarpa Kiely, with particular reference to its epiphytology and control at Letaba. D.Sc. (Agric.) thesis. [Google Scholar]
- Kotzé J.M. Epidemiology and control of citrus black spot in South Africa. Plant Disease Reporter. 1981;65:945–950. doi: 10.1094/PDIS.1997.81.8.851. [DOI] [PubMed] [Google Scholar]
- Kotzé J.M. History and epidemiology of citrus black spot in South Africa. Proceedings of the International Society of Citriculture. 1996;2:1296–1299. [Google Scholar]
- Kotzé J.M. Black spot. In: Timmer L.W., Garnsey S.M., Graham J.H., editors. Compendium of Citrus Diseases. 2nd edn. The American Phytopathological Society; St. Paul, MN, USA: 2000. pp. 23–25. [Google Scholar]
- Lee Y.S., Huang C.S. Effect of climatic factors on the development and discharge of ascospores of the citrus black spot fungus. Journal of Taiwan Agricultural Research. 1973;22:135–144. [Google Scholar]
- Leggieri M.C., Decontardi S., Bertuzzi T. Modeling growth and toxin production of toxigenic fungi signaled in cheese under different temperature and water activity regimes. Toxins. 2017;9:1–17. doi: 10.3390/toxins9010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.K., Phookamsak R., Doilom M. Towards a natural classification of Botryosphaeriales. Fungal Diversity. 2012;57:149–210. [Google Scholar]
- Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Mabberley D.J. Citrus (Rutaceae): a review of recent advances in etymology, systematics and medical applications. Blumea. 2004;49:481–498. [Google Scholar]
- Magarey R.D., Hong S.C., Fourie P.H. Prediction of Phyllosticta citricarpa using an hourly infection model and validation with prevalence data from South Africa and Australia. Crop Protection. 2015;75:104–114. [Google Scholar]
- McOnie K.C. Source of inoculum of Guignardia citricarpa, the Citrus Black Spot pathogen. Phytopathology. 1964;54:64–67. [Google Scholar]
- McOnie K.C. Germination and infection of citrus by ascospores of Guignardia citricarpa in relation to control of black spot. Phytopathology. 1967;57:743–746. [Google Scholar]
- Meyer L., Sanders G.M., Jacobs R. A one-day sensitive method to detect and distinguish between the citrus black spot pathogen Guignardia citricarpa and the endophyte Guignardia mangiferae. Plant Disease. 2006;90:97–101. doi: 10.1094/PD-90-0097. [DOI] [PubMed] [Google Scholar]
- Miles A.K., Tan Y.P., Tan M.K. Phyllosticta spp. on cultivated Citrus in Australia. Australasian Plant Pathology. 2013;42(4):461–467. [Google Scholar]
- Miles A.K., Willingham S.L., Cooke A.W. Field evaluation of strobilurins and a plant activator for the control of citrus black spot. Australasian Plant Pathology. 2004;33:371–378. [Google Scholar]
- Moncalvo J.M., Wang H.H., Hseu R.S. Phylogenetic relationships in Ganoderma inferred from the internal transcribed spacer and 25S ribosomal DNA sequences. Mycologia. 1995;87:223–238. [Google Scholar]
- Morton J.F. Creative Resource Systems, Inc.; Miami, Florida, USA: 1987. Fruits of warm climates; pp. 160–168. [Google Scholar]
- Myllys L., Stenroos S., Thell A. New genes for phylogenetic studies of lichenized fungi: glyceraldehyde-3-phosphate dehydrogenase and beta-tubulin genes. Lichenologist. 2002;34:237–246. [Google Scholar]
- Nei M. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolosi E. Origin and taxonomy. In: Khan I.A., editor. Citrus genetics, breeding and biotechnology. CAB International; Oxfordshire UK: 2007. pp. 19–43. [Google Scholar]
- Noronha M.D.A. Universidade de Sao Paulo; 2002. Escala diagramatica para avaliacao da mancha preta em folhas de citros efeito da temperatura e da duracao do molhamento na prepenetracao de conidios de Guignardia citricarpa Kiely [Phyllosticta citricarpa (McAlp.) Van der Aa] Agronomy: Concentration of Plant Pathology Masters Dissertation. [Google Scholar]
- Nylander J.A.A. Evolutionary Biology Centre, Uppsala University; 2004. MrModeltest v. 2. Program distributed by the author. [Google Scholar]
- O'Donnell K., Kistler H.C., Cigelnik E. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okane I., Lumyong S., Nakagiri A. Extensive host range of an endophytic fungus, Guignardia endophyllicola (anamorph: Phyllosticta capitalensis) Mycoscience. 2003;44:353–363. [Google Scholar]
- Okane I., Nakagiri A., Ito T. Identity of Guignardia sp. inhabiting ericaceous plants. Canadian Journal of Botany. 2001;79:101–109. [Google Scholar]
- Paul I., van Jaarsveld A.S., Korsten L. The potential global geographical distribution of Citrus Black Spot caused by Guignardia citricarpa (Kiely): likelihood of disease establishment in the European Union. Crop Protection. 2005;24:297–308. [Google Scholar]
- Peakall R., Smouse P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perryman S.A.M., Clark S.J., West J.S. Splash dispersal of Phyllosticta citricarpa conidia from infected citrus fruit. Scientific Reports. 2014;4:6568. doi: 10.1038/srep06568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persoon C.H. Belin-Leprieur; Paris, France: 1818. Traité sur les champignons comestibles, contenant l'indication des espèces nuisibles; a l'histoire des champignons. [Google Scholar]
- Petrak F. Über die Gattungen Guignardia Viala & Ravaz und Discosphaerina v. Höhnel. Sydowia. 1957;11:435–445. [Google Scholar]
- Pu J., Xie Y., Zhang X. Preinfection behaviour of Phyllosticta musarum on banana leaves. Australasian Plant Pathology. 2008;37:60–64. [Google Scholar]
- Rakotoniriana E.F., Munaut F., Decock C. Endophytic fungi from leaves of Centella asiatica: occurrence and potential interactions within leaves. Antonie van Leeuwenhoek. 2008;93:27–36. doi: 10.1007/s10482-007-9176-0. [DOI] [PubMed] [Google Scholar]
- Ramón-Laca L. The introduction of cultivated Citrus to Europe via northern Africa and the Iberian Peninsula. Economic Botany. 2003;57:502–514. [Google Scholar]
- Rayner R.W. CMI and British Mycological Society; Kew, Surrey, England: 1970. A mycological colour chart. [Google Scholar]
- Reis R.F., Timmer L.W., de Goes A. Effect of temperature, leaf wetness and rainfall on the production of Guignardia citricarpa ascospores and on black spot severity on sweet orange. Fitopatologia Brasileira. 2006;31:29–34. [Google Scholar]
- Rodrigues K.F., Samuels G.J. Fungal endophytes of Spondias mombin leaves in Brazil. Journal of Basic Microbiology. 1999;39:131–135. [Google Scholar]
- Rodrigues K.F., Sieber T.N., Grünig C.R. Characterization of Guignardia mangiferae isolated from tropical plants based on morphology, ISSR-PCR amplifications and ITS1-5.8S-ITS2 sequences. Mycological Research. 2004;108:45–52. doi: 10.1017/s0953756203008840. [DOI] [PubMed] [Google Scholar]
- Romão A.S., Spósito M.B., Andreote F.D. Enzymatic differences between the endophyte Guignardia mangiferae (Botryosphaeriaceae) and the citrus pathogen G. citricarpa. Genetics and Molecular Research. 2011;10:243–252. doi: 10.4238/vol10-1gmr952. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Denis M., Guarnaccia V., Polizzi G. Symptomatic Citrus trees reveal a new pathogenic lineage in Fusarium and two new Neocosmospora species. Persoonia. 2018;40:1–25. doi: 10.3767/persoonia.2018.40.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch C.L., Shoemaker R.A., Seifert K.A. A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia. 2006;98:1041–1052. doi: 10.3852/mycologia.98.6.1041. [DOI] [PubMed] [Google Scholar]
- Schubert T.S., Dewdney M.M., Peres N.A. First report of Guignardia citricarpa associated with citrus black spot on sweet orange [Citrus sinensis (L.) Osbeck] in North America. Plant Disease. 2012;96:1225. doi: 10.1094/PDIS-01-12-0101-PDN. [DOI] [PubMed] [Google Scholar]
- Schubert T.S., Sutton B., Jeyaprakash A. Pest Alert DACS-P-01723. Florida Department of Agriculture and Consumer Services, Division of Plant Industry; Gainesville, FL, USA: 2010. Citrus black spot (Guignardia citricarpa) discovered in Florida. [Google Scholar]
- Schutte G.C., Kotze C., van Zyl J.G. Assessment of retention and persistence of copper fungicides on orange fruit and leaves using fluorometry and copper residue analyses. Crop Protection. 2012;42:1–9. [Google Scholar]
- Schutte G.C., Mansfield R.I., Smith H. Application of azoxystrobin for control of benomyl-resistant Guignardia citricarpa on ‘Valencia’ oranges in South Africa. Plant Disease. 2003;87:784–788. doi: 10.1094/PDIS.2003.87.7.784. [DOI] [PubMed] [Google Scholar]
- Smith H., Wingfield M.J., Crous P.W. Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. South African Journal of Botany. 1996;62:86–88. [Google Scholar]
- Snowdon A.L. Wolfe Scientific Ltd.; London, UK: 1990. A colour atlas of post-harvest diseases and disorders of fruits and vegetables, vol. 1: general introduction and fruits; pp. 62–63. [Google Scholar]
- Spósito M.B., Amorim L., Bassanezi R.B. Spatial pattern of black spot incidence within citrus trees related to disease severity and pathogen dispersal. Plant Pathology. 2008;57:103–108. [Google Scholar]
- Sposito M.B., Amorim L., Bassanezi R.B. Relative importance of inoculum sources of Guignardia citricarpa on the citrus black spot epidemic in Brazil. Crop Protection. 2011;30:1546–1552. [Google Scholar]
- Su Y.Y., Cai L. Polyphasic characterisation of three new Phyllosticta spp. Persoonia. 2012;28:76–84. doi: 10.3767/003158512X645334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan A., Johnston P.R., Park D. Two new pathogenic ascomycetes in Guignardia and Rosenscheldiella on New Zealand's pygmy mistletoes (Korthalsella: Viscaceae) Studies in Mycology. 2011;68:237–247. doi: 10.3114/sim.2011.68.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung G.H., Sung J.M., Hywel-Jones N.L. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized incongruence using a combinational bootstrap approach. Molecular Phylogenetics and Evolution. 2007;44:1204–1223. doi: 10.1016/j.ympev.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, Massachusetts: 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods), v. 4.0b10. [Google Scholar]
- Tamura K., Stecher G., Peterson D. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongkantha S., Lumyong S., McKenzie E.H.C. Fungal saprobes and pathogens occurrence on tissues of Dracaena loureiri and Pandanus spp. Fungal Diversity. 2008;30:149–179. [Google Scholar]
- Tran N.T., Miles A., Dietzgen R.G. Sexual reproduction in the Citrus Black Spot pathogen, Phyllosticta citricarpa. Phytopathology. 2017 doi: 10.1094/PHYTO-11-16-0419-R. [DOI] [PubMed] [Google Scholar]
- Truter M. University of Pretoria; Pretoria, South Africa: 2010. Epidemiology of citrus black spot disease in South Africa and its impact on phytosanitary trade restrictions. PhD thesis. [Google Scholar]
- USDA APHIS (United States Department of Agriculture Animal and Plant Health Inspection Service) Center for Plant Health Science and Technology, Plant Epidemiology and Risk Analysis Laboratory; Raleigh, NC, USA: 2010. Risk assessment of Citrus spp. fruit as a pathway for the introduction of Guignardia citricarpa Kiely, the organism that causes Citrus Black Spot disease. [Google Scholar]
- Viala P., Ravaz L. Sur la dénomination botanique (Guignardia bidwellii) du black-rot. Bulletin de la Société Mycologique de France. 1892;8:63. [Google Scholar]
- Vicent A., Armengol J., García-Jiménez J. Rain fastness and persistence of fungicides for control of Alternaria brown spot of citrus. Plant Disease. 2007;91:393–399. doi: 10.1094/PDIS-91-4-0393. [DOI] [PubMed] [Google Scholar]
- Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen G., Huang F. Phyllosticta species associated with citrus diseases in China. Fungal Diversity. 2012;52:209–224. [Google Scholar]
- Wang N.Y., Zhang K., Huguet-Tapia J.C. Mating type and simple sequence repeat markers indicate a clonal population of Phyllosticta citricarpa in Florida. Phytopathology. 2016;106:1300–1310. doi: 10.1094/PHYTO-12-15-0316-R. [DOI] [PubMed] [Google Scholar]
- White T.J., Bruns T., Lee S. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR protocols: a guide to methods and applications. Academic Press; San Diego, California: 1990. pp. 315–322. [Google Scholar]
- Whiteside J.O. Sources of inoculum of the black spot fungus, Guignardia citricarpa, in infected Rhodesian orchards. Rhodesia, Zambia and Malawi Journal of Agricultural Research. 1967;5:171–177. [Google Scholar]
- Wicht B., Petrini O., Jermini M. Molecular, proteomic and morphological characterization of the ascomycete Guignardia bidwellii, a phylogenetic re-evaluation of Phyllosticta (Botryosphaeriales) agent of grape black rot: a polyphasic approach to fungal identification. Mycologia. 2012;104:1036–1045. doi: 10.3852/11-242. [DOI] [PubMed] [Google Scholar]
- Wikee S., Lombard L., Nakashima C. A phylogenetic re-evaluation of Phyllosticta (Botryosphaeriales) Studies in Mycology. 2013;76:1–29. doi: 10.3114/sim0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikee S., Lombard L., Crous P.W. Phyllosticta capitalensis, a widespread endophyte of plants. Fungal Diversity. 2013;60:91–105. [Google Scholar]
- Wikee S., Udayanga D., Crous P.W. Phyllosticta: an overview of current status of species recognition. Fungal Diversity. 2011;51:43–61. [Google Scholar]
- Wingfield M.J., de Beer Z.W., Slippers B. One fungus, one name promotes progressive plant pathology. Molecular Plant Pathology. 2012;13:604–613. doi: 10.1111/j.1364-3703.2011.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M.H., Crous P.W., Henderson J. Phyllosticta species associated with freckle disease of banana. Fungal Diversity. 2012;56:173–187. [Google Scholar]
- Wulandari N.F., To-anun C., Hyde K.D. Phyllosticta citriasiana sp. nov., the cause of Citrus tan spot of Citrus maxima in Asia. Fungal Diversity. 2009;34:23–39. [Google Scholar]
- Yonow T., Hattingh V., de Villiers M. CLIMEX modelling of the potential global distribution of the citrus black spot disease caused by Guignardia citricarpa and the risk posed to Europe. Crop Protection. 2013;44:18–28. [Google Scholar]
- Yuan Z., Chen Y., Yang Y. Diverse non-mycorrhizal fungal endophytes inhabiting an epiphytic, medicinal orchid (Dendrobium nobile): estimation and characterization. World Journal of Microbiology and Biotechnology. 2009;25:295–303. [Google Scholar]
- Zavala M.G.M., Er H.L., Goss E.M. Genetic variation among Phyllosticta strains isolated from citrus in Florida that are pathogenic or nonpathogenic to citrus. Tropical Plant Pathology. 2014;39:119–128. [Google Scholar]
- Zhang K., Wang N.Y., Dewdney M.M. APS Annual Meeting, August 1–5, Pasadena, California, USA. 2015. Lonely peninsula: the mating-type and population of Phyllosticta citricarpa in Florida. [Google Scholar]
- Zhou N., Chen Q., Carroll G. Polyphasic characterization of four new plant pathogenic Phyllosticta species from China, Japan, and the United States. Fungal Biology. 2015;119:433–446. doi: 10.1016/j.funbio.2014.08.006. [DOI] [PubMed] [Google Scholar]