Fig. 2.
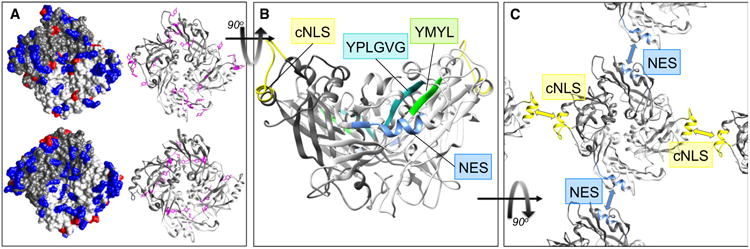
Homology-modeled structure and oligomerization of NiV matrix. (A) Surface features of M proteins that enable membrane association. Homology-modeled structure of dimeric NiV-M protein (top) generated using SWISS-MODEL [133–135] based on the existing X-ray crystal structure of NDV-M protein (bottom), PDB ID code 4G1G [25]. The two monomers are shaded light gray and dark gray in both the space-filling models (left), and the ribbon diagrams (right). The structures are oriented to show the surface that interacts with the cellular plasma membrane, with features thought to contribute to membrane association highlighted: There is an enrichment of surface-exposed positively charged residues (blue, left) and a planar distribution of tyrosine residues (fushia, right). (B) Sequences involved in NiV-M nuclear-cytoplasmic trafficking. Homology-modeled NiV-M (as in A) viewed from the side, with the membrane-binding surface at the top. The cNLS (yellow) and NES (blue) [33] are both located in surface-exposed α-helices. The putative late domain sequences ‘YMYL’ [74] (green) and ‘YPLGVG’ [107] (cyan) form a contiguous motif in the predicted tertiary structure, and together comprise two strands of the β-sheet that underlies the NES helix. (C) The cNLS and NES sequences form the two predicted dimer–dimer interfaces in NiV-M. Homology-modeled NiV-M (as A) shaded as (B), oriented to show the membrane-interacting surface. The blue and yellow arrows indicate the two dimer–dimer interfaces as identified for NDV-M [25], which are contributed by the NES (blue) and cNLS (yellow) α-helices, respectively.
