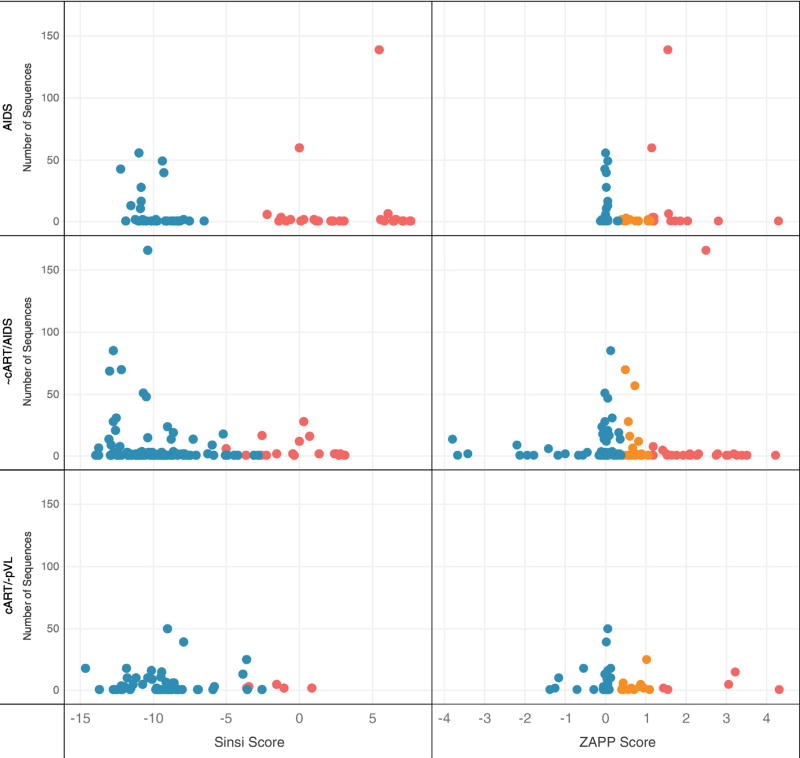
Fig. 2.
Coreceptor usage prediction for WebPSSM sinsi (left) and ZAPP (right) algorithms. 556 brain and 1249 non-brain sequences were evaluated from 26 different tissue types (71 tissues in total) collected from 15 subjects. Sequences from subjects were grouped into three categories (y-axis): No cART/AIDS (934 sequences), variable cART/AIDS (536 sequences), and cART+ subjects who were virally suppressed (335 sequences). Predicted X4 variants (red), predicted dual-tropic variants (R5X4) (orange) and predicted R5 variants (blue) are indicated. WebPSSM predicts envelope sequences as R5 (x < −6.96) or X4 (x > −2.88); for sequences with scores that are intermediate between R5 or X4, the program uses the “11/25 rule,” which proposes that a positively charged amino acid at positions 11 or 25 is likely an X4 isolate. ZAPP predicts over a continuous range using thresholds for coreceptor assignment as R5 (x < 0.378), R5X4 (0.378 ≤ x < 1.13), and X4 (x ≥ 1.13). The results demonstrate no remarkable differences among coreceptor affinity pre- and post-ART. Many of the autopsy samples derived from cART+/virally suppressed subjects revealed various tissue-based pathologies (Brew and Chan, 2014). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)