Abstract
Background
Studies on gut microbiota regarding colorectal carcinogenesis, including sessile serrated adenoma (SSA), have been scarce. The aim of this study is to investigate the role of mucosa-associated gut microbiota in the colorectal carcinogenesis.
Methods
We collected biopsy samples of normal rectal mucosa during colonoscopy from healthy control and patients with conventional adenoma, SSA, and colorectal cancer (CRC), respectively (n = 6). Pyrosequencing for 16S rRNA gene of bacteria was performed to compare gut microbiota.
Results
The most abundant phylum in total samples was Proteobacteria (55.6%), followed by Firmicutes (27.4%) and Bacteroidetes (11.6%). There was no significant difference in relative abundance of the phylum level among the four groups. Fusobacterium nucleatum, known to be frequently detected during colorectal carcinogenesis, was found in only one sample of patient with SSA. The rarefaction curves showed that the diversity of mucosal communities of patients with CRC is the lowest among the four groups and the diversity of mucosal communities of patients with SSA is higher than that of healthy control. Among the four groups, Shannon’s and Simpson’s index for diversity was the lowest and the highest in the patients with CRC, respectively; it did not reach statistical significance. The proportion of genus Pseudomonas was very high in the samples of patients with stage II–IV CRC compared with those with stage I CRC (59.3% vs. 0.3%, P = 0.064).
Conclusions
Our study suggests no significant role of mucosa-associated gut microbiota in the colorectal carcinogenesis. Further study for many samples or using fecal material could be helpful.
Keywords: Microbiota, Colonic neoplasms, Colon cancer, Colon adenoma, Sessile serrated adenoma
INTRODUCTION
Chronic inflammation following chronic infection is one of main mechanism in human carcinogenesis. Development of gastric cancer after long-time Helicobacter pylori infection is a typical example.1 The role of single pathogen in colorectal carcinogenesis was also studied for a long time. Fusobacterium nucleatum, Escherichia coli, Enterococcus faecalis, Streptococcus gallolyticus, and enterotoxigenic Bacteroides fragilis are candidate microorganisms that were pointed out as initial triggers in the development of colorectal cancer (CRC). It was reported that these bacteria are related with CRC in epidemiology and the possibility as causative microorganism in colorectal carcinogenesis was suggested by several animal models.2 However, none of these bacteria succeeded to prove definite casual relation with CRC. At present, no study showed that elimination of these organisms in human prevents CRC.
Recently, rapid development of new generation sequencing methods in the field of microorganism made it possible to identify gut microbiota that was unlikely to be culturable. In addition, these new technologies paved the path for analysis of massive genetic data of gut microbiota: metagenomics. Given this background, instead of single pathogen, the change in overall gut microbiota, interaction between each microbiota, and metabolites that bacteria produce are actively being studied in the field of colorectal carcinogenesis. It is well known that CRC usually develops through serial mutations after adenomatous change rather than de novo genesis.3 Therefore, researches regarding change of gut microbiota in adenomatous polyps that are precancerous lesions are also actively being carried out. Compared to healthy control, in the patients with adenomatous polyp and CRC, gut dysbiosis such as decreased diversity of gut microbiota is frequently observed.4 Increase of Fusobacterium and decrease of butyrate-producing bacteria are also suggested. Considering field cancerization, that is an important concept in the carcinogenesis,5 we can infer that these changes in gut microbiota would already occur not only in the neoplastic lesions but also in surrounding normal-looking mucosa. Actually, it was reported that gut microbiota of colorectal adenoma and near-by normal mucosa are similar to each other; but they are different from that of healthy control.6 In addition, Fusobacterium increased already in the normal rectal mucosa of patients with adenomatous polyps compared to that of healthy control.7
Recently, sessile serrated adenoma (SSA) has received attention as a main cause of right colon cancer and interval CRC. SSA is known to progress to CRC through different pathway from conventional adenomatous polyp.8 Therefore, we can expect that the role and change of gut microbiota would be different from that in conventional adenomatous polyp. However, studies on gut microbiota regarding colorectal carcinogenesis including SSA have been scarce.
Gut microbiota can be largely divided into two groups depending on their anatomical localization: fecal-luminal microbiota and mucosa-associated microbiota.9 Fecal-luminal microbiota occupies the lumen of the intestine. It can be acquired relatively easily by collecting stool. Therefore, most large-scale studies regarding human gut microbiota including key studies, such as MeTaHIT cohort and Human Microbiome Project analyzed fecal-luminal microbiota.10,11 However, fecal-luminal microbiota is more transient and it is more affected by diet than mucosa-associated microbiota. Mucosa-associated microbiota resides on the surface of the intestinal epithelium. Because it is more consistent and interacts with host cells more directly and intimately than fecal-luminal microbiota, it is considered more closely related with CRC.4,9 Nevertheless, to acquire it, endoscopic biopsy which is invasive procedure is required and bowel preparation for colonoscopy can change mucosa-associated microbiota.12 Therefore, studies for gut microbiota and CRC often used fecal-luminal microbiota rather than mucosa-associated microbiota.
From this background, the aim of this study is to compare mucosa-associated gut microbiota among healthy control, patients with conventional adenoma, SSA, and CRC.
MATERIALS AND METHODS
1. Subjects and tissue preparation
We collected biopsy samples of normal rectal mucosa during colonoscopy from four different groups: healthy control, patients with conventional adenoma, SSA and CRC, respectively. Rectal mucosal tissues of healthy control were acquired from samples that were collected for other study (IRB No. B-1308/215-008). Healthy control was defined as subject who underwent screening colonoscopy for the first time of life and in whom the result of colonoscopy was normal. Tissues of subjects with SSA were also acquired from samples that were collected for another study (IRB No. B-1204/152-004). For the present study, subjects who had no synchronous adenomatous polyps among subjects with SSA were selected. Tissues of subjects with conventional adenoma and subjects with CRC were selected from tissue pools of professor NK. All subjects with adenomatous polyp who enrolled in the present study had advanced colorectal neoplasms defined as adenomatous polyp larger than 1 cm, with villous portion, or with high grade dysplasia. Sex and age within 3 years were matched for each group. We confirmed that there was no history of antibiotics administration within three months before the day of mucosal biopsy in all subjects. All mucosal samples were obtained from normal proximal rectum (at approximately 18 cm from anal verge) by colonoscopic biopsy. The material was centrifuged at 20,000 ×g for 30 minutes to pellet bacteria. The pellet was processed to extract total microbial DNA using a commercial kit (iNtRON Biotechnology, Seongnam, Korea). This study was approved by the Institutional Review Board of the Seoul National University Hospital (IRB No. B-1606-352-302).
2. Barcoded 454-pyrosequencing of the 16S rRNA gene
DNA samples from four groups were subjected to pyrosequencing. PCR amplification was performed using primers targeting the V1 to V3 regions of the 16S rRNA gene from extracted DNA. For bacterial amplification, barcoded primers of 9F (5′-CCTATCCCCTGTGTGCCTTGGCAGTC-TCAG-AC-AGAGTTTGATCMTGGCTCAG-3′; underlined sequence indicates the target region primer) and 541R (5′-CCATCTCATCCCTGCGTGTCTCCGAC-TCAG-X-AC-ATTACCGCGGCTGCTGG-3′; ‘X’ indicates the unique barcode for each subject) (http://oklbb.ezbiocloud.net/content/1001) were used. Amplifications were carried out using an initial denaturation at 95°C for 5 minute, followed by 30 cycles of denaturation at 94°C for 30 seconds, primer annealing at 55°C for 45 seconds, and extension at 72°C for 90 seconds, with a final elongation at 72°C for 5 minutes. PCR products were analyzed by 2% agarose gel electrophoresis and visualized using a Gel Doc system (BioRad, Hercules, CA, USA). The amplified products were purified with the QIAquick PCR purification kit (Qiagen, Valencia, CA, USA). Equal concentrations of purified products were pooled together, and short fragments (non-target products) were removed with Ampure beads kit (Agencourt Bioscience, Beverly, MA, USA). The quality and product size were assessed on a Bioanalyzer 2100 (Agilent, Palo Alto, CA, USA) using a DNA 7500 chip. Mixed amplicons were subjected to emulsion PCR, and then deposited on Picotiter plates. The sequencing was carried out at Chunlab Inc. (Seoul, Korea) with GS Junior Sequencing system, the modified laboratory benchtop form of 454 sequencing systems (Roche, Branford, CT, USA) according to the manufacturer’s instructions.
3. Pyrosequencing data analysis
Reads obtained from different samples were sorted by unique barcodes of each PCR product. The barcode, linker, and primer sequences were removed from the original sequencing reads. Reads containing two or more ambiguous nucleotides, reads with a low quality score (average score < 25), or reads shorter than 300 bp, were discarded. Potential chimeric sequences were detected by the Bellerophon method, which compares the BLASTN search results between the forward half and reverse half sequences.13 After removing the chimeric sequences, the taxonomic classification of each read was assigned against the EzTaxon-e database (http://eztaxon-e.ezbiocloud.net),14 which contains the 16S rRNA gene sequence of type strains that have valid published names and representative species level phylotypes of either cultured or uncultured entries in the GenBank database with complete hierarchical taxonomic classification from the phylum to the species.
The richness and diversity of samples were determined by abundance-based coverage estimators (ACE), Chao1 and Jackknife estimation. In addition, Simpson diversity index and Shannon diversity index at 3% distance were calculated in the CL community program (Chunlab Inc.). Good’s method was used to calculate sequencing coverage.15
Random subsampling was conducted to equalize the read size of samples for comparing different read sizes among samples. To compare the operational taxonomic units (OTUs) between samples, shared OTUs were obtained with the XOR analysis of the CL community program (Chunlab Inc.). UniFrac-based principal coordinates analysis was used to compare the microbial communities of sub-groups.
4. Statistical analysis
PASW for Windows ver. 18.0 (IBM Co., Armonk, NY, USA) was used for the statistical analysis. Because the data was not normally distributed (as demonstrated by the Shapiro-Wilk test), quantitative variables among four groups were compared using a Kruskal-Wallis test. Post hoc comparisons of pairwise differences between two groups were evaluated by the Mann-Whitney U-test using the modified Bonferroni procedure for multiple comparison adjustment. All results were considered statistically significant when P-values were less than 0.05.
RESULTS
1. Relative abundance of mucosal associated microbiota
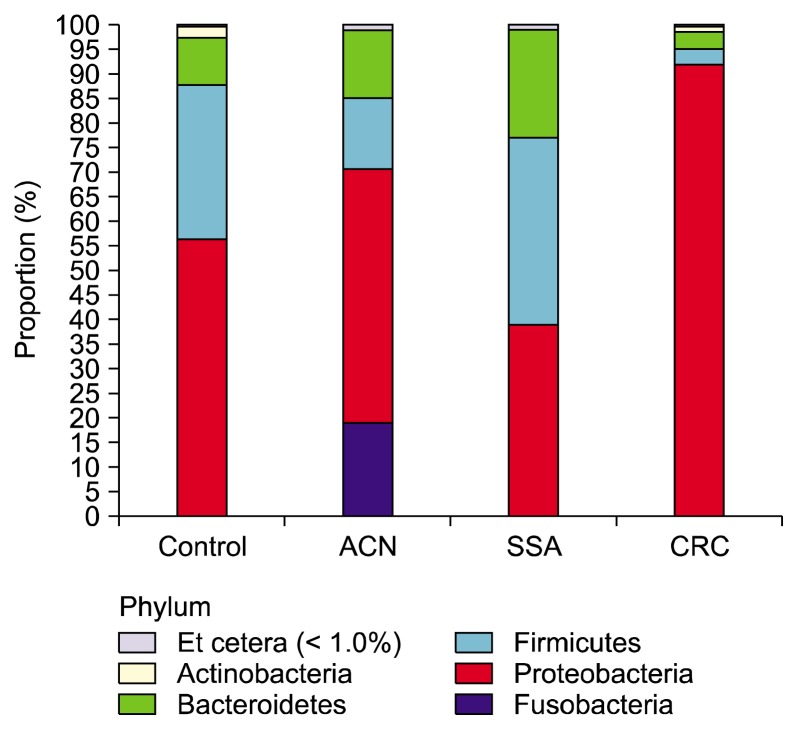
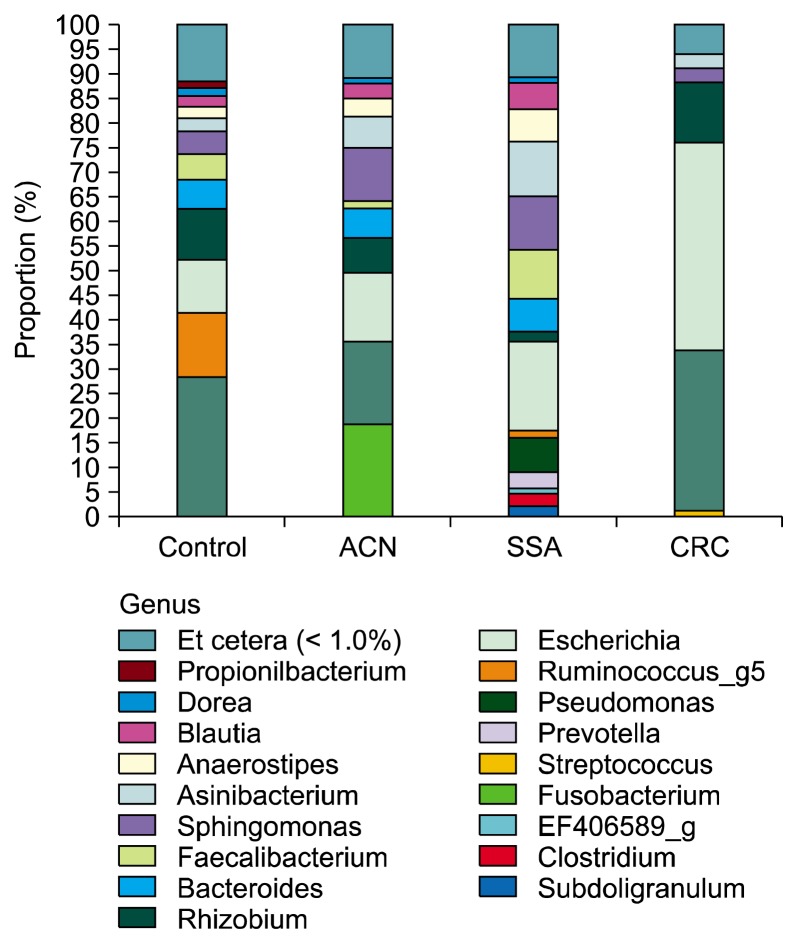
Total 24 subjects were enrolled in the present study: three subjects by each sex in four groups. There was no significant difference in the mean age among the four groups. (mean, 61.75 ± 10.86, P = 0.791). The basal characteristics of pyrosequencing data analysis and species richness indicators in the rectal mucosa samples are shown in Table 1. The mean number of total and valid 16S rRNA reads for each sample was 6,070 and 3,022, respectively. The most abundant phylum in total samples was Proteobacteria (55.6%), followed by Firmicutes (27.4%), Bacteroidetes (11.6%), Fusobacteria (3.2%), and Actinobacteria (1.7%). Figure 1 shows relative abundance of the phylum level in the rectal mucosa samples by pyrosequencing. There was no significant difference in relative abundance of most abundant phyla among the four groups (P > 0.05). Relative abundance of microbiota among the four groups was not significantly different in the genus level (Fig. 2). The most abundant species in total samples was E. coli (24.6%), followed by Pseudomonas cremoricolorata (21.7%) and Rhizobium leguminosarum (8.0%). F. nucleatum, which is known to be frequently detected in patients with CRC, was found in only one sample of patient with SSA. The percentage of F. nucleatum sequence was also very low in that sample (0.04%). The abundance of Faecalibacterium prausnitzii and Roseburia hominis that are principle ones of butyrate-producing bacteria16 were not different among the four groups.
Table 1.
The basal characteristics of pyrosequencing data analysis and species richness indicators in the rectal mucosa samples
| Characteristic | Control (n = 6) | ACN (n = 6) | SSA (n = 6) | CRC (n = 6) | P-value |
|---|---|---|---|---|---|
| Valid reads (median) | 1,435 | 3,374 | 2,557 | 3,929 | 0.551 |
| OTUs (median) | 131 | 128 | 80 | 101 | 0.684 |
| Good’s library coverage (%) | 96.0 | 97.3 | 95.9 | 98.8 | 0.470 |
| Diversity index (median) | |||||
| ACE | 259.89 | 171.45 | 143.04 | 174.92 | 0.433 |
| Chao1 | 208.20 | 166.21 | 119.06 | 158.90 | 0.795 |
| Jackknife | 233.44 | 173.11 | 125.94 | 181.95 | 0.825 |
| Shannon | 3.16 | 3.39 | 2.75 | 2.79 | 0.343 |
| Simpson | 0.10 | 0.06 | 0.14 | 0.16 | 0.584 |
ACN, advanced colorectal neoplasm; SSA, sessile serrated adenoma; CRC, colorectal cancer; OTU, operational taxonomic units; ACE, abundance-based coverage estimators.
Figure 1.
Relative abundance of the phylum level (% similarity) in the rectal mucosa samples by pyrosequencing. ACN, advanced colorectal neoplasm; SSA, sessile serrated adenoma; CRC, colorectal cancer.
Figure 2.
Relative abundance of the genus level (% similarity) in the rectal mucosa samples by pyrosequencing. ACN, advanced color-ectal neoplasm; SSA, sessile serrated adenoma; CRC, colorectal cancer.
2. Alpha-diversity
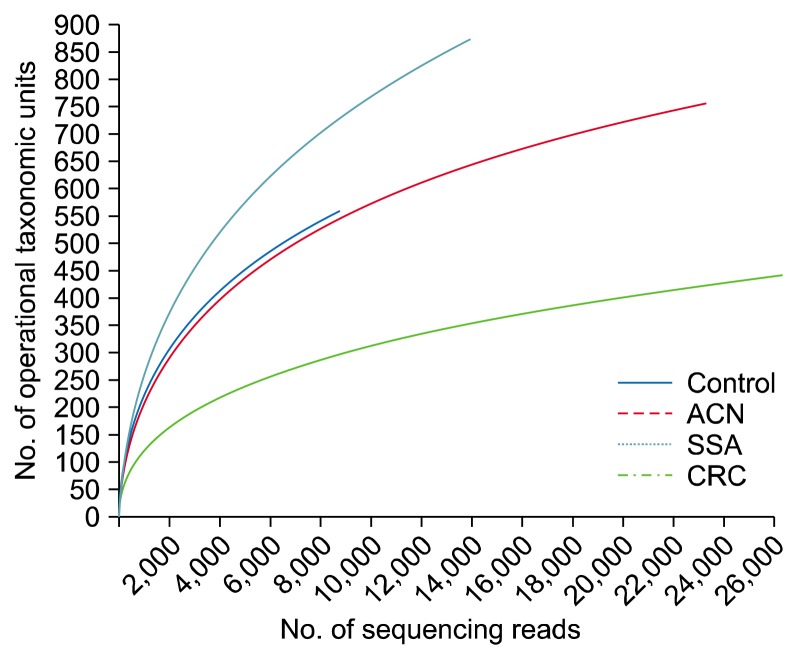
The overall Good’s coverage was 96.1%. Figure 3 shows rarefaction curve for the comparison of the microbial communities in the rectal mucosa samples. The rarefaction curves shows that the diversity of mucosal communities of subjects with CRC is the lowest among the four groups and the diversity of mucosal communities of subjects with SSA is higher than that of healthy control. Species richness indicators such as ACE, Chao1, and Jackknife were not different among the four groups (Table 1). Shannon’s index for diversity was the lowest and Simpson’s index for diversity was the highest in the subjects with CRC among the four groups, although they did not reach statistical significance.
Figure 3.
Rarefaction curve for the comparison of the microbial communities in the rectal mucosa samples by pyrosequencing. ACN, advanced colorectal neoplasm; SSA, sessile serrated adenoma; CRC, colorectal cancer.
3. Beta-diversity
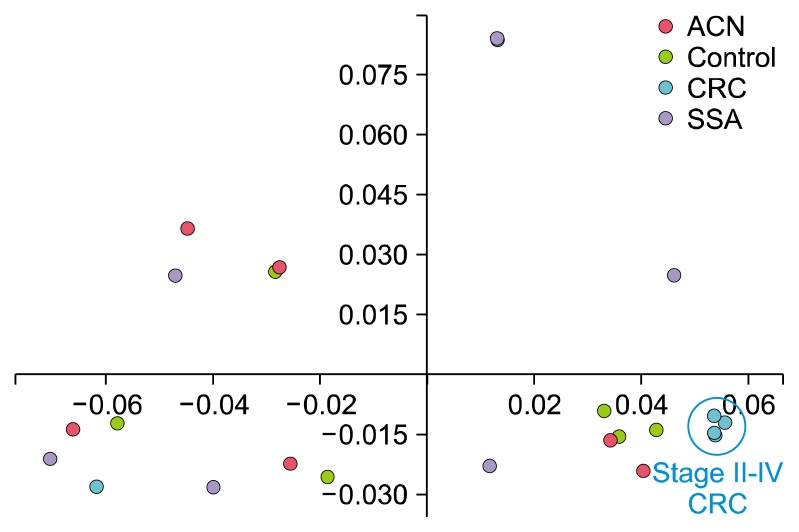
Figure 4 shows un-weighted UniFrac-based principal coordinates analysis of the microbial communities in the rectal mucosa. The samples were not definitely separated according to their groups. However, four samples from CRC patients with stage II–IV were gathered very closely and definitely separated from two samples from patients with stage I CRC; the clinicopathologic data of six subjects of CRC group are summarized in Table 2. Although it failed to reach statistical significance due to the small sample size, the proportion of genus Pseudomonas was very high in the samples of patients with stage II–IV CRC compared in the samples of patients with stage I CRC (59.3% vs. 0.3%, P = 0.064).
Figure 4.
Un-weighted UniFrac-based principal coordinates analysis of the microbial communities in the rectal mucosa. This analysis is based on taxa clustered at 97% sequence 16S rRNA gene identity. Samples obtained from patients of control, advanced colorectal neoplasm, sessile serrated adenoma, and colorectal cancer are represented by yellow-green, pink, purple and cyan circles, respectively. ACN, advanced colorectal neoplasm; CRC, colorectal cancer; SSA, sessile serrated adenoma.
Table 2.
Clinicopathologic data of six patients with colorectal cancer
| Sample name | Sex | Age (yr) | Location of cancer | TNM classification | Stagea |
|---|---|---|---|---|---|
| CC 7 | F | 50 | Rectum | T3N1bM0 | IIIB |
| CC 8 | M | 58 | Rectum | T3N0M0 | IIA |
| CC 9 | F | 55 | S-colon | T2N0M0 | I |
| CC 10 | M | 55 | Rectum | T3N2M1 | IV |
| CC 11 | F | 78 | S-colon | T2N0M0 | I |
| CC 12 | M | 79 | Cecum | T3N2bM0 | IIIC |
F, female; M, male; S, sigmoid.
Stage was defined by the American Joint Committee on Cancer Cancer Staging Manual, 7th edition (2010).
DISCUSSION
When there is change in the gut microbiota in patient with colorectal neoplasm, it is very difficult to determine whether it is cause or consequence of colorectal neoplasm. Tjalsma et al.17 suggested bacterial driver-passenger model for CRC. They reported that change of gut microbiota in the neoplastic tissue is useful to determine passenger bacteria and change of gut microbiota in non-neoplastic tissue is rather helpful to decide driver bacteria. Based on this study, we analyzed gut microbiota of normal-looking mucosa of the subjects. Overall results showed that there was no significant difference in mucosa-associated gut microbiota among healthy control, patients with conventional adenoma, SSA, and CRC. However, this finding is likely related to low power stemming from small sample size. In the rarefaction curve, we found tendency of decreasing diversity of gut microbiota from healthy control to advanced colorectal neoplasm and CRC group. This finding corresponds well with the results of earlier studies which reported that diversity of mucosa-associated microbiota of patients with conventional adenoma or CRC decreased compared to that of healthy control.18,19 On the contrary, gut microbiota of SSA group showed higher diversity than that of healthy control in the rarefaction curve. This result implies that the change of gut microbiota in patients with SSA differs from change during conventional adenoma-carcinoma sequence. Similarly to our results, Peters et al.20 reported that SSA did not differ in diversity or composition from controls and few taxa were differentially abundant between SSA cases and controls. They suggested that gut microbiota may play a role in colorectal carcinogenesis through the development of conventional adenoma only. Our finding compensates Peters et al.’ study20 which has limitation in that it is derived from analysis of fecal-luminal microbiota.
In a large study regarding premalignant lesions and CRC including SSA, Ito et al.21 reported that F. nucleatum was identified in premalignant colorectal lesions regardless of histopathology. Because this study used qPCR for identification of F. nucleatum instead of metagenomics, it is inappropriate to compare it directly with our study. Nevertheless, F. nucleatum was observed in a very low frequency in the samples of our study and we speculated this due to small sample size.
Definite separation of gut microbiota according to each group was not found in PCoA analysis in the present study. However, interestingly, gut microbiota of patients with stage II or higher CRC was separated from that of patients with stage I CRC. Specifically, the proportion of genus Pseudomonas was very high in the samples of patients with stage II or high CRC. Because we collected mucosal samples of CRC patients before the start of treatment, such as surgery or chemotherapy, we speculated that Pseudomonas might be passenger bacteria which rapidly increase after change of intestinal milieu as CRC progresses. However, we could not find any article supporting our finding. Instead, it was reported that Enterobacteriaceae which is a branch of Proteobacteria same as Pseudomonas and contains genera, such as E. coli, Klebsiella spp., and Proteus spp. blooms in the inflamed microenvironment in the gut.22 Further studies using many samples are required for the investigating the role of Pseudomonas during colorectal carcinogenesis.
This study has several limitations. It is well known that the proportion of Proteobacteria is higher in mucosa-associated microbiota than in fecal-luminal microbiota.23 Nevertheless, the proportion of Proteobacteria in all groups is excessively high compared to other studies.24 Considering that Proteobacteria is ubiquitous bacteria in the ecosystem,25 we cannot exclude the possibility that the samples were contaminated at some stage. Therefore, we reviewed whole process from tissue sampling to bacterial DNA sequencing; but we could not find any problematic steps in particular. In addition, because the sample size was too small for each group, it is very difficult to make a definite conclusion regarding gut microbiota during colorectal carcinogenesis based on the results of this study alone.
Our study suggests no significant role of mucosa-associated gut microbiota in the colorectal carcinogenesis. Further study for many samples or using fecal material could be helpful.
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (NRF) grant for the Global Core Research Center (GCRC) funded by the Korea government (MSIP) (No. 2011-0030001).
Footnotes
CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Correa P, Houghton J. Carcinogenesis of Helicobacter pylori. Gastroenterology 2007;133:659–72. 10.1053/j.gastro.2007.06.026 [DOI] [PubMed] [Google Scholar]
- 2.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe 2014;15:317–28. 10.1016/j.chom.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies RJ, Miller R, Coleman N. Colorectal cancer screening: prospects for molecular stool analysis. Nat Rev Cancer 2005;5:199–209. 10.1038/nrc1569 [DOI] [PubMed] [Google Scholar]
- 4.Dulal S, Keku TO. Gut microbiome and colorectal adenomas. Cancer J 2014;20:225–31. 10.1097/PPO.0000000000000050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato T, Suzuki K, Muto Y, Sasaki J, Tsujinaka S, Kawamura YJ, et al. Multiple primary malignancies involving primary sporadic colorectal cancer in Japan: incidence of gastric cancer with colorectal cancer patients may be higher than previously recognized. World J Surg Oncol 2015;13:23. 10.1186/s12957-014-0432-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu Y, Chen J, Zheng J, Hu G, Wang J, Huang C, et al. Mucosal adherent bacterial dysbiosis in patients with colorectal adenomas. Sci Rep 2016;6:26337. 10.1038/srep26337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PLoS One 2013;8:e53653. 10.1371/journal.pone.0053653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leggett B, Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology 2010;138:2088–100. 10.1053/j.gastro.2009.12.066 [DOI] [PubMed] [Google Scholar]
- 9.Fung TC, Artis D, Sonnenberg GF. Anatomical localization of commensal bacteria in immune cell homeostasis and disease. Immunol Rev 2014;260:35–49. 10.1111/imr.12186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59–65. 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–14. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tyler AD, Smith MI, Silverberg MS. Analyzing the human microbiome: a “how to” guide for physicians. Am J Gastroenterol 2014;109:983–93. 10.1038/ajg.2014.73 [DOI] [PubMed] [Google Scholar]
- 13.Huber T, Faulkner G, Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 2004;20:2317–9. 10.1093/bioinformatics/bth226 [DOI] [PubMed] [Google Scholar]
- 14.Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 2012;62:716–21. 10.1099/ijs.0.038075-0 [DOI] [PubMed] [Google Scholar]
- 15.Li XX, Wong GL, To KF, Wong VW, Lai LH, Chow DK, et al. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS One 2009;4:e7985. 10.1371/journal.pone.0007985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014;63:1275–83. 10.1136/gutjnl-2013-304833 [DOI] [PubMed] [Google Scholar]
- 17.Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol 2012;10:575–82. 10.1038/nrmicro2819 [DOI] [PubMed] [Google Scholar]
- 18.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One 2012;7:e39743. 10.1371/journal.pone.0039743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst 2013;105:1907–11. 10.1093/jnci/djt300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters BA, Dominianni C, Shapiro JA, Church TR, Wu J, Miller G, et al. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome 2016;4:69. 10.1186/s40168-016-0218-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer 2015;137:1258–68. 10.1002/ijc.29488 [DOI] [PubMed] [Google Scholar]
- 22.Zeng MY, Inohara N, Nuñez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol 2017;10:18–26. 10.1038/mi.2016.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ringel Y, Maharshak N, Ringel-Kulka T, Wolber EA, Sartor RB, Carroll IM. High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut Microbes 2015;6:173–81. 10.1080/19490976.2015.1044711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durbán A, Abellán JJ, Jiménez-Hernández N, Ponce M, Ponce J, Sala T, et al. Assessing gut microbial diversity from feces and rectal mucosa. Microb Ecol 2011;61:123–33. 10.1007/s00248-010-9738-y [DOI] [PubMed] [Google Scholar]
- 25.Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol 2015;33:496–503. 10.1016/j.tibtech.2015.06.011 [DOI] [PubMed] [Google Scholar]