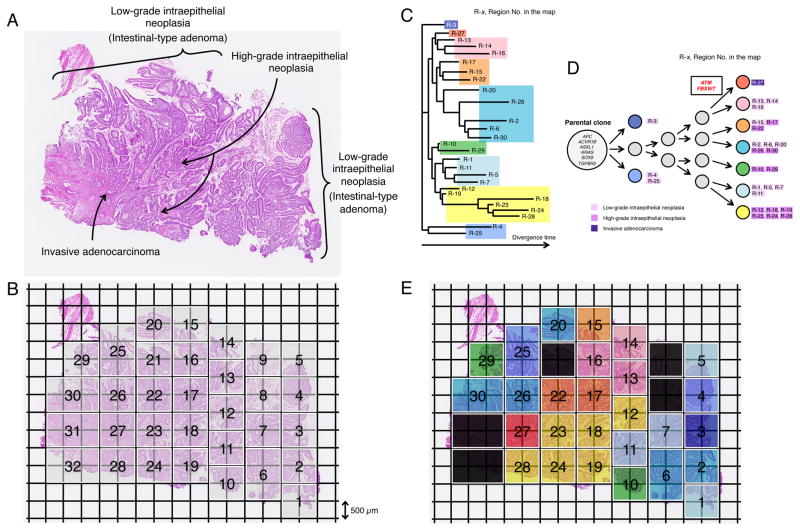
Figure 5. Geographic Mapping of Subclones Based on Multi-region Exome Sequencing and Proposed Clonal Evolution of an Ampullary Carinoma with Low-grade and High-grade Intraepithelial Neoplasias.
(A) Histology of a sample consisting of the components of low-grade intraepithelial neoplasia (Intestinal-type adenoma), high-grade intraepithelial neoplasia and invasive carcinoma (hematoxylin and eosin stain).
(B) A whole section was broken up into 32 squares of 1 mm2 units (thickness, 18 μm) by Glass Chip Macrodissection (GCM) technology and was dissected. Genomic DNA (gDNA) was individually extracted from the same compartment using the five serial sections and subjected to Agilent SureSelect Human All Exon v4.0 based hybrid selection followed by exome library construction for Illumina sequencing.
(C) Phylogenetic tree was drawn based on the sequencing data of synonymous mutations.
(D) Proposed clonal evolution model based on the phylogenetic tree during tumor progression. Cancer driver genes reported in COSMIC are represented. The mutations of the cancer driver genes, ATM and FBXW7 are observed only in region No.27 where cancer cells start to be invasive morphologically. Gray circles indicate predicted subclones based on the phylogenetic tree.
(E) Sections are colored, corresponding to the colors of the phylogenetic tree in (C) and (D).
See also Table S11.