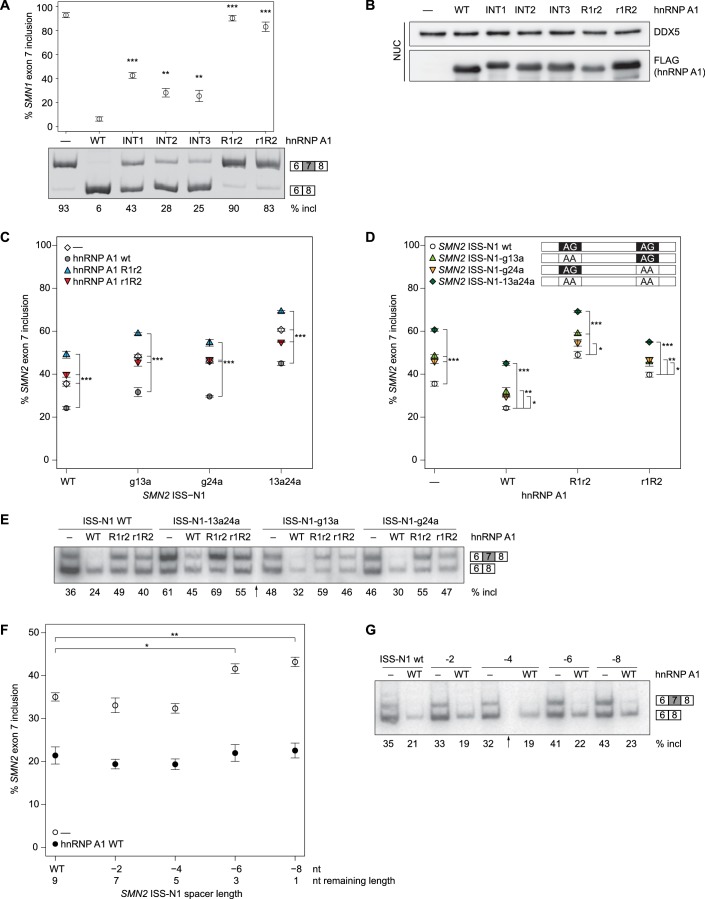
Figure 5. SMN splicing repression depends on hnRNP A1 organization and the bipartite ISS-N1 motif.
(A) Data from alternative splicing assay in HEK293T cells. Top panel, quantification of exon 7 inclusion in SMN1 upon overexpression of wild-type or mutant hnRNP A1. Bottom panel, PAGE with PCR products used for quantification. (B) Western Blot of nuclear extracts used in A. (C, D) Quantification of exon 7 inclusion in SMN2 in relation to overexpression of wild-type or mutant hnRNP A1 (C) or to mutation of the hnRNP A1 binding sites in the ISS-N1 (D). Both figures (C) and (D) contain are different representations of the same data. (E) Radioautography of PAGE with PCR products used for quantification in (C, D).The separation between two individual gels is indicated by an arrow. (F) Quantification of exon 7 inclusion in SMN2 in relation to the spacer length in-between the two hnRNP A1 binding sites within the ISS-N1. The wild-type ISS-N1 has a spacer length of 9 nt. (G) Radioautography of PAGE with PCR products used for quantification in (F). The separation between two individual gels is indicated by an arrow. Data information: All data points represent the mean of the biological replicates. Error bars correspond to the S.E.M. (B) **p=0.00113 (INT2), p=0.00360 (INT3), ***p<0.001; (C) ***p<0.001; (D) *p<0.5, **p<0.01, ***p<0.001; (F) *p=0.01176, **p=0.00287. All tested with one-way ANOVA (WT = 0), for panel (C, D) SMN2 wt co-transfections n = 4, otherwise n = 3. The mean of the replicate is always given below the corresponding PAGE for a better readability. If not otherwise indicated 0.5 µg hnRNP A1 were co-transfected with 1 µg of the mini-gene.
DOI: http://dx.doi.org/10.7554/eLife.25736.022
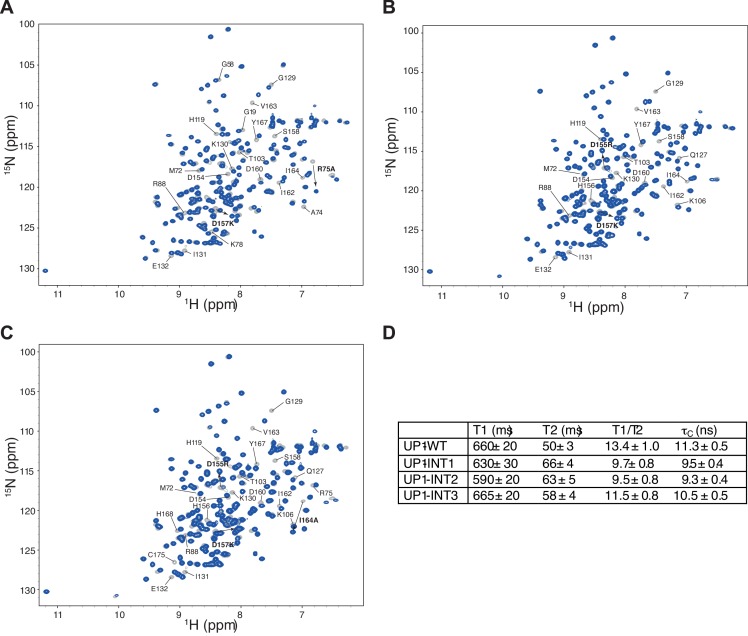
Figure 5—figure supplement 1. Protein mutants disrupt the inter-RRM interface.
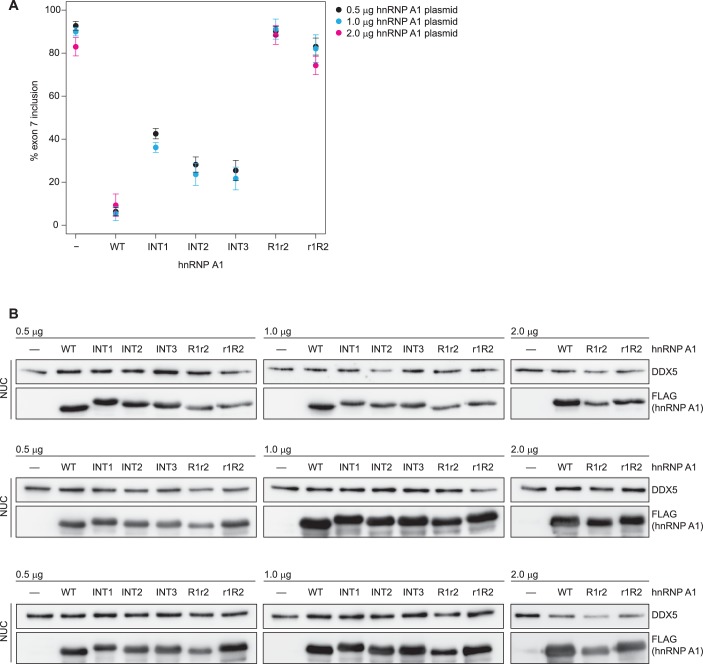
Figure 5—figure supplement 2. No dose dependency of hnRNP A1 overexpression.
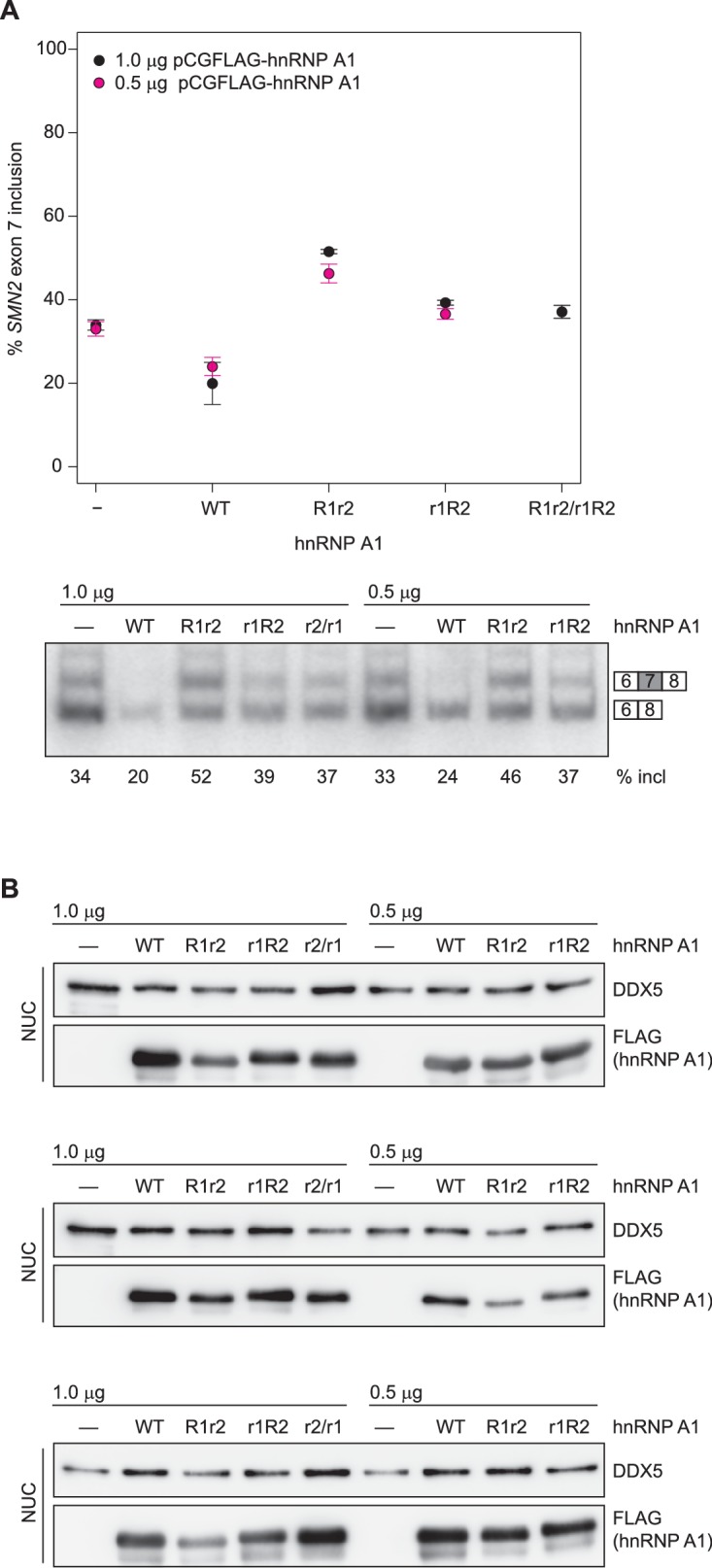
Figure 5—figure supplement 3. Western blots for SMN2 exon 7 splicing experiments in HEK293T cells.
Figure 5—figure supplement 4. hnRNP A1 RRM mutants can not complement each other in SMN2 splicing assay.