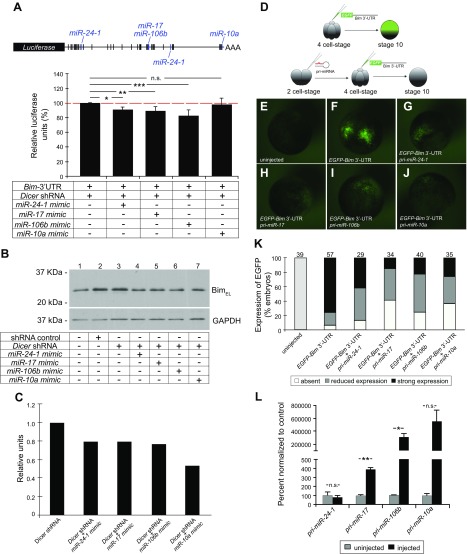
Figure 6.
miR-24-1, miR-17, miR-106b, and miR-10a repress the expression of Bim in vitro and in vivo. A) Schematic of the pmiRGLO-Bim 3′-UTR luciferase reporter vector containing the mouse Bim 3′-UTR. Potential miRNA binding sites predicted by in silico analyses are represented by vertical bars. Blue: predicted binding sites for miR-24-1, miR-17, miR-106b, and miR-10a. Luciferase reporter assays performed in HEK 293 cells transfected with pmiRGLO-Bim 3′-UTR in the presence of Dicer shRNA and miRNA mimics (miR-24-1, miR-17, miR-106b, and miR-10a). There was a reduction of ∼10, 11, and 18% in luciferase activity in the presence of miR-24-1, miR-17, and miR-106b, respectively. No significant change in luciferase activity was observed in the presence of miR-10a mimic (n = 3 independent experiments). Error bars ± sem. N.s., nonsignificant. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (1-way ANOVA with Turkey’s post hoc test). B) Western blot analysis comparing Bim levels in protein extracts of HEK 293 cells transfected with either shRNA control or Dicer shRNA, in the presence of miRNA mimics. The miR-24-1, miR-17, miR-106b, and miR-10a mimics decreased the expression of BimEL. GAPDH was used as a loading control. C) Densitometric analyses of the bands on the Western blot shown in (B). The relative levels of BimEL were calculated by normalizing with GAPDH. D–K) Xenopus laevis embryos were microinjected with a synthetic full-length Bim 3′-UTR fused to the EGFP reporter gene, in the presence or absence of pri-mmu-miR-24-1, pri-mmu-miR-17, pri-mmu-miR-106b, or pri-mmu-miR-10a. D) EGFP expression was assessed at stage 10. E–K) Representative images (E–J) and the quantification (K) of 3 independent experiments are shown. The total number of embryos analyzed is indicated above the individual bars. L) qPCR analysis demonstrating the levels of mature miRNAs (mmu-miR-24-1, mmu-miR-17, mmu-miR-106b, and mmu-miR-10a) in embryos injected with the primary transcripts, pri-mmu-miR-24-1, pri-mmu-miR-17, pri-mmu-miR-106b, and pri-mmu-miR-10a (n = 3 biologic replicates per condition from 3 independent experiments. Each replicate represented 4 embryos that were pooled). Error bars ± sem. N.s., nonsignificant. *P ≤ 0.05, **P ≤ 0.01 (Student’s unpaired Student’s t test).