Figure 1.
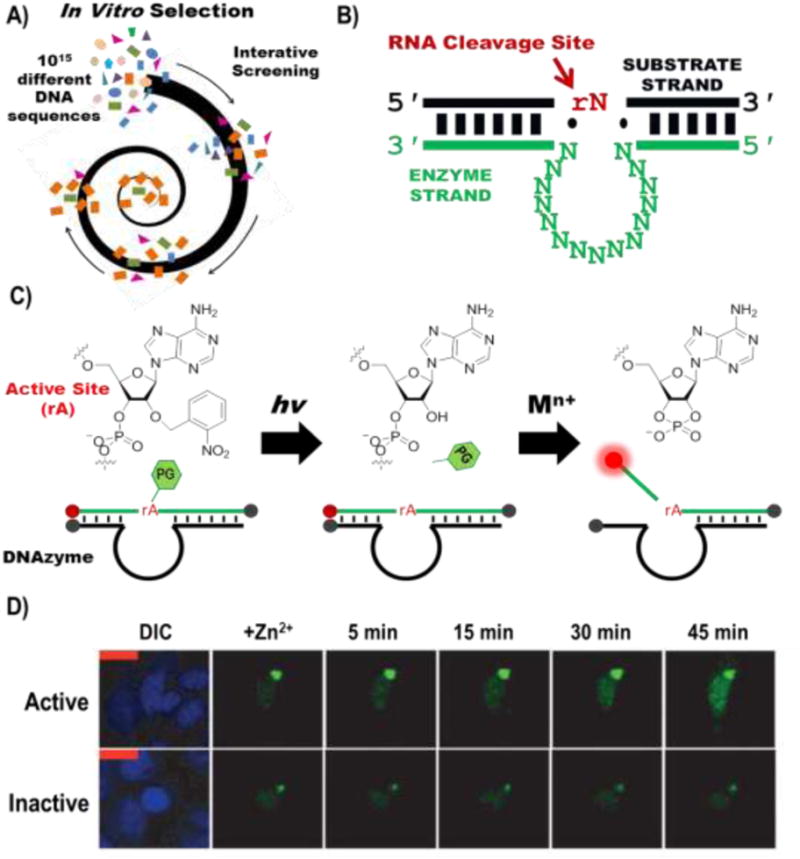
DNAzymes are selected via an iterative combinatorial selection strategy called in vitro selection (A). The resulting DNAzyme (B) has binding arms which hybridize according to Watson-Crick base pairing, indicated by the black and green bars, which are highly programmable, an enzymatic region, shown as repeated N, where N is A, C, G or T, and an RNA cleavage site indicated in red at rN, where N can be A, C, G, T or U. (C) By functionalizing the DNAzyme with fluorophores (red) and quenchers (grey), it can be turned into a catalytic beacon, which has a turn-on fluorescent signal in the presence of a selective monovalent, divalent, or trivalent metal-ion cofactor. The catalytic beacon can be caged using the photolabile nitrobenzyl group on the 2′-OH. An example of the efficacy of the photocaged catalytic beacon is shown in D (figure modified from ref. 49)
