Figure 4.
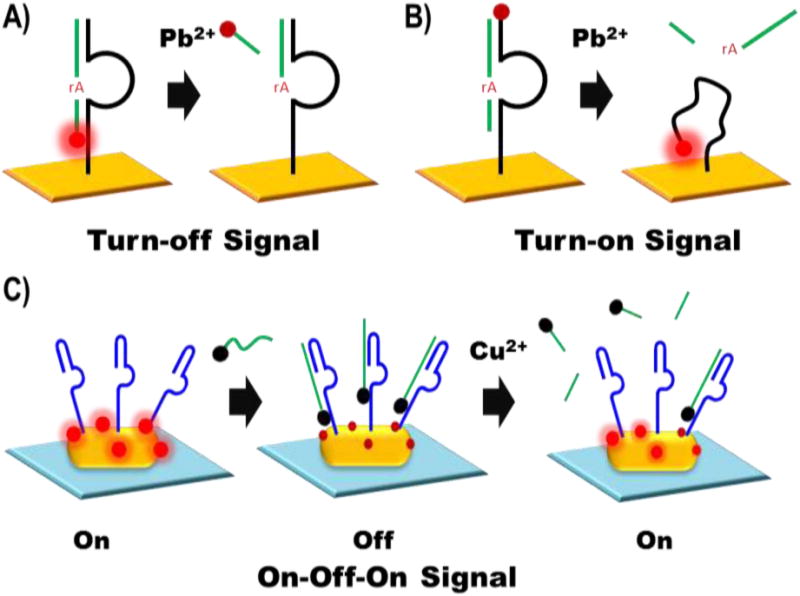
Schematics of the turn-off (A) turn-on (B) and “On-Off-On” signal (C) DNAzyme sensor constructs. The turn-off sensor (A) uses a DNAzyme enzyme sequence (black) tethered to an electrode (gold surface) to cleave a DNAzyme substrate sequence (green) at the RNA active site (indicated by rA in red). The cleavage reaction will release the Ru(bpy)3-NHS tag (shown in bright red when “on” and dark red when “off”) into solution turning “off” the luminescence signal. The turn-on sensor uses release of the DNAzyme substrate to allow the Ru(bpy)3-NHS labeled enzyme sequence to bend towards the electrode and generate a turn-on signal. The “On-Off-On” sensor (C) initially has a gold nanorod (yellow oval) coated in 3,4,9,10-perylenetetracarboxylic acid (indicated in bright red when excited and dark red when quenched) and the enzyme sequence of the Cu2+-dependent DNAzyme (dark blue). The introduction of a substrate strand (green) attached to a ferrocene quencher (black circle) turns off the signal and the subsequent introduction of Cu2+ cause the cleavage and subsequent release of the quencher to regenerate a turn-on signal.
