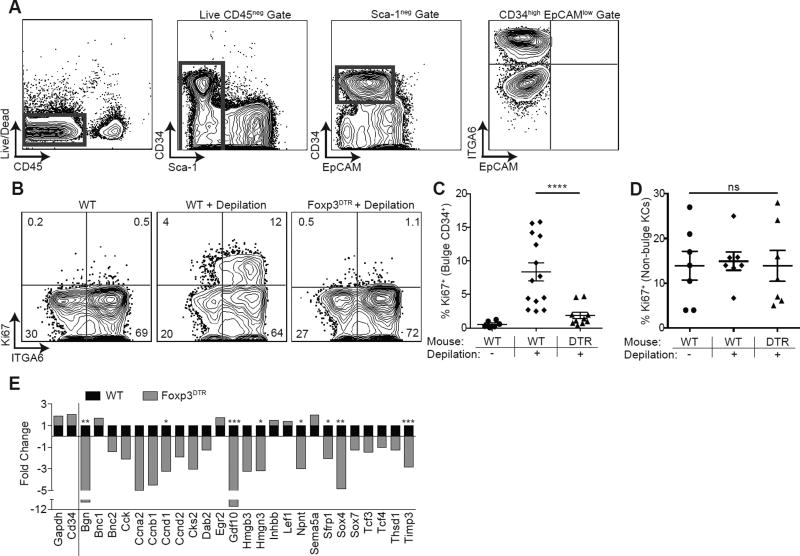
Figure 4. Tregs are required for HFSC proliferation and differentiation.
(A) Flow cytometric gating strategy to identify CD34+ integrin α6high (ITGA6) bulge HFSCs. Foxp3DTR mice or control mice were treated with DT on days −2, −1, depilated on day 0 to induce anagen and DT administered again on days 1 and 3 (i.e., early regimen). (B) Representative flow cytometric plots of Ki67 expression in bulge HFSCs between WT and FoxpDTR mice 4 days after depilation. (C) Flow cytometric quantification of Ki67+ bulge HFSCs and (D) non-bulge keratinocytes 4 days after depilation. RNA sequencing was performed on FACS purified bulge HFSCs at day 4 post-depilation from control (WT) or Treg depleted (Foxp3DTR) mice. (E) Fold change in HFSC differentiation genes in WT and FoxpDTR mice, expressed as fold change relative to WT (where a value of 1 = no change). Genes to the left of the solid line represent control genes. Significance values are calculated based on transcript expression level. Data are mean ± s.e.m. *P<0.05, **P<0.01 ***P<0.001, **** P<0.0001, ns = no significant difference, One-way ANOVA (C and D); RNA-Seq Differential Expression analysis, WT vs. Foxp3DTR for each respective gene (E). See also Figure S4.