Figure 1.
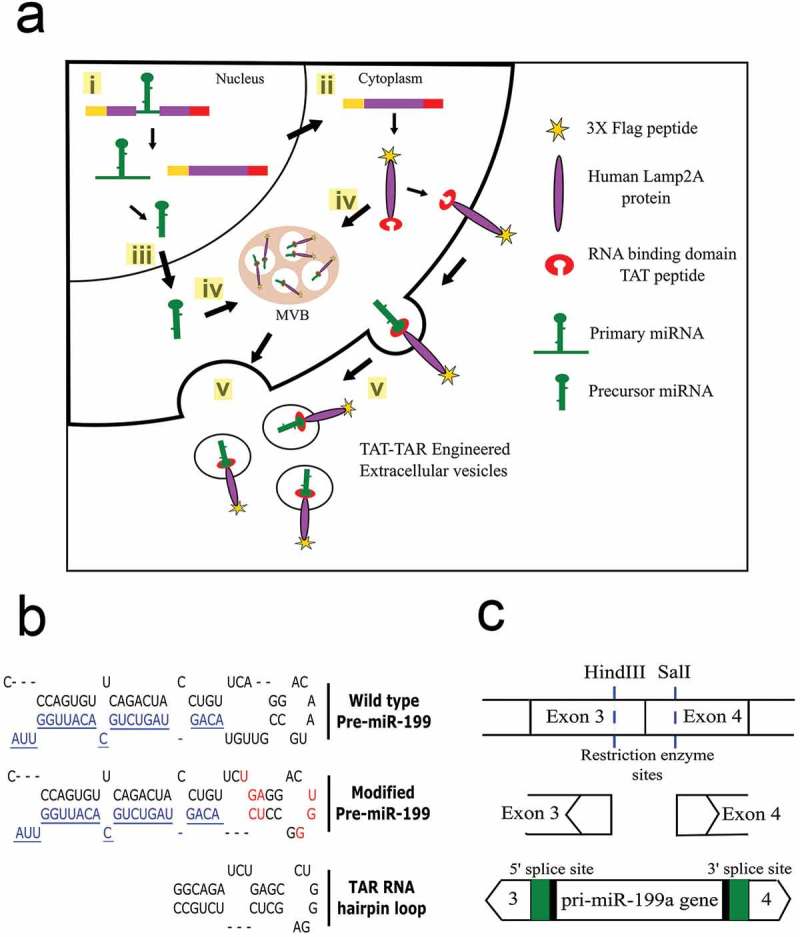
Overview of strategy to load cargo RNAs into EVs. (a) Sequence of events leading to loading of cargo RNA into the EVs. (i) The modified miR-199a gene engineered into an intron of the targeting fusion protein gene is spliced from the mRNA in the nucleus of the HEK293T cells. (ii) The fusion protein is translated from the spliced mRNA (iii) while the modified pri-miR-199a is processed to mature miR-199a-3p. (iv) Both the targeting, fusion protein and pre-miR-199a are directed into exosomes or microvesicles; the targeting protein through alignment of the Lamp2a transmembrane domain and the pre-miRNA-199a through binding of the modified precursor to the TAT peptide on the luminal C-terminus of the targeting protein. (v) EVs are shed into the extracellular space and collected by ultracentrifugation. (b) Unique restriction sites were located in exons 3 and 4 of the Lamp2a cDNA. The 370 bp chimeric intron containing the pri-miR-199a gene, 5′ and 3′ flanking sequences of exons 3 and 4, respectively and the 5′ and 3′ splice sites were cloned into the restriction sites of the targeting vector. Green colour depicts the consensus sequences that were engineered to increase the splicing efficiency. (c) Sequence of the wild type pri-miR-199a sequence (top) with the mature miR-199a-3p coloured blue. Eight nt contained within the loop and stem portions (red) were modified in the pri-miR-199a sequence (middle) to resemble that portion of the TAR RNA that binds to the TAT protein (bottom).
