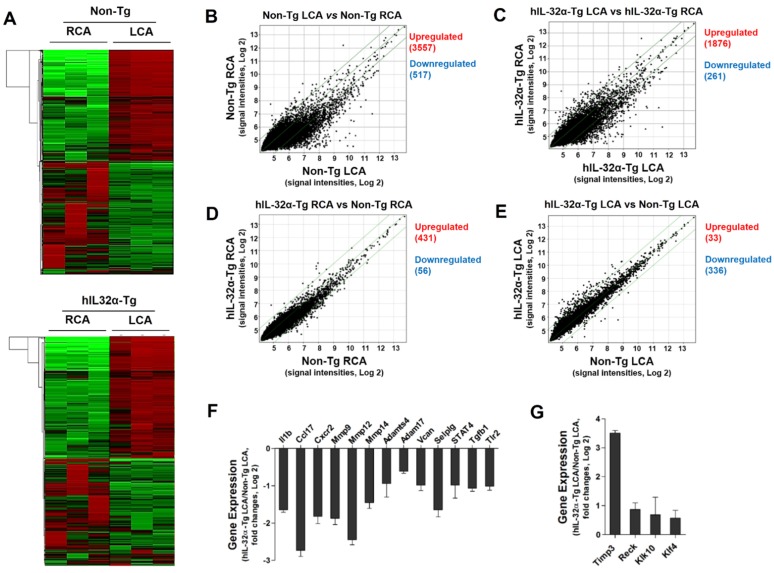
Figure 5.
Global gene expression profiles in the carotid artery endothelium of hIL-32α-Tg mice. Total RNAs were obtained from the intima of mouse LCA and RCA at 48 h after ligation. Affymetrix GeneChip® Mouse Gene 2.0 ST arrays containing 770,317 mouse gene probes were used. (A) Hierarchical clustering analysis results indicating the genes whose expression significantly differed between hIL-32α-Tg mice LCA endothelium and non-Tg mice LCA endothelium are shown as heat maps. Each column represents a single sample pooled from 3 different LCAs, and each row represents a single gene probe. (B-E) Scatter plots show normalized intensities of each probe under 4 comparison conditions: (B) non-Tg LCA vs. non-Tg RCA, (C) hIL-32α-Tg LCA vs. hIL-32α-Tg RCA, (D) hIL-32α-Tg RCA vs. non-Tg RCA, and (E) hIL-32α-Tg LCA vs. non-Tg LCA after ligation. Genes are shown that were up-regulated (red) or down-regulated (blue) (≥2-fold difference) at a false discovery rate of ≤10% in LCA compared with RCA. Microarray results for selected (F) pro-atherogenic and (G) anti-atherogenic genes are shown as the fold-increase or fold-decrease of gene expression in hIL-32α-Tg LCA compared with that in non-Tg LCA on a log2 scale (mean ± SEM). All genes shown in the graphs were significantly different between groups (n = 3, p < 0.05 as determined by Student's t-test).