Abstract
Venoms are a rich source for the discovery of molecules with biotechnological applications, but their analysis is challenging even for state-of-the-art proteomics. Here we report on a large-scale proteomic assessment of the venom of Loxosceles intermedia, the so-called brown spider. Venom was extracted from 200 spiders and fractioned into two aliquots relative to a 10 kDa cutoff mass. Each of these was further fractioned and digested with trypsin (4 h), trypsin (18 h), pepsin (18 h), and chymotrypsin (18 h), then analyzed by MudPIT on an LTQ-Orbitrap XL ETD mass spectrometer fragmenting precursors by CID, HCD, and ETD. Aliquots of undigested samples were also analyzed. Our experimental design allowed us to apply spectral networks, thus enabling us to obtain meta-contig assemblies, and consequently de novo sequencing of practically complete proteins, culminating in a deep proteome assessment of the venom. Data are available via ProteomeXchange, with identifier PXD005523.
Keywords:
Subject terms: Proteomics, Protein analysis, Entomology
Background & Summary
Scientists have long enlisted venoms in their quest to characterize novel molecules with biotechnological applications1,2. The literature provides innumerous examples of venom-derived applications, ranging from biopesticides to medical applications. In particular, works on serpent venom are, unarguably, success stories. Some examples are: Batroxobin, a widely used thrombin-like enzyme and commonly extracted from the venom of Bothrops atrox and Bothrops moojeni, has been used as a replacement for thrombin in bleeding injuries3; Ecarin, from Echis carinatus, as the primary reagent for laboratorial tests that monitor anticoagulation4; and Captopril, developed from peptides of the Bothrops jararaca venom, as a widely adopted inhibitor of the angiotensin converting enzyme (ACE). Other examples of venom-derived drugs include: Aggrastat, for myocardial infarct and ischemia; Ancrod, for stroke; Defibrase, for acute cerebral infarction and angina pectoris; Exanta, used as an anti-coagulant; Hemocoagulase, for hemorrhage; and Integrilin, for acute coronary syndrome5. Venoms have also been used to search for inhibitors derived from other species (e.g., Didelphis marsupialis)6,7.
Motivated by all the successful research on snake venoms, efforts have been geared towards spider toxins. In particular, those from the Loxosceles genus are already being used in at least four general application fronts, viz.: as therapeutic anti-venom sera8; as tools in molecular and cellular biology research; and as aids in drug development and production of selective and environmentally friendly bioinsecticides5. Peptides originating from the venom of Thrixopelma pruriens have been used in the treatment of pain and inflammation9; the T×2–5 and T×2–6 neuropeptides from the Phoneutria nigriventer venom, for treating erectile dysfunctions10; and distinct bioactive peptides from spider venoms, in the treatment of diverse diseases, such as cancer11. Taken together, toxins have served as an endless treasure trove for biotechnological applications.
Spider venoms, in particular, comprising mainly proteins and peptides2,5,12,13 and displaying great diversity in their toxins, have drawn considerable attention. Yet, characterizing venoms poses great challenges even for state-of-the-art proteomic strategies: in fact, most species lack a reference sequence genome14 and the post-translational modifications of venoms vary greatly. Moreover, current mainstream strategies are not tailored towards performing de novo sequencing of the large (i.e., greater than tryptic), biologically active peptides that abound in venoms. Indeed, peptide-centric approaches are oblivious to whether a sequenced peptide originates from a larger peptide or a full protein, but obtaining the complete sequence of these larger molecules will undoubtedly fuel a great diversity of biotechnological applications. In this regard, it is our view that widely adopted proteomic strategies such as peptide spectrum matching (PSM)15,16 and mainstream de novo sequencing17 only reveal the tip of the iceberg in terms of what can be unveiled from venoms.
One of our goals has been to characterize the venom of the so-called brown spiders (the Loxosceles genus). Altogether, their venom is composed of a complex cocktail of biologically active compounds, with toxins ranging up to 40 kDa and over18. To the best of our knowledge, an in-depth, comprehensive proteomic profiling of the Loxosceles venom tailored towards the discovery of new molecules has so far remained elusive. Currently, there are several descriptions of enzymatic and non-enzymatic proteins from distinct Loxosceles species19,20. In 2003, a study aimed to investigate whether venoms of phylogenetically-related groups of Haplogyne spiders possess sphingomyelinase-D (SMD) toxins21. The study included 10 Loxosceles species and 2 Sicarius species, among other spider genera. The Amplex Red Phospholipase-D assay kit indicated SMD activity and these results were further supported by a Surface-Enhanced Laser Desorption/Ionization (SELDI) Time-of-Flight (TOF) analysis showing mass spectral peaks with m/z’s corresponding to those of SMD. Loxosceles SMDs, later referred to as phospholipases-D (PLDs), are known to be the major component of Loxosceles venoms and are the most well characterized toxin family in brown spider venoms. In 2005, two-dimensional protein profiles of the L. intermedia, L. laeta, and L. gaucho venoms were determined, but protein identification was focused only on the SMD toxins of the L. gaucho venom22. The identification of seven spots of interest was first attempted using data from Matrix-Assisted Laser Desorption/Ionization (MALDI) Time-of-Flight (TOF) Mass Spectrometry (MS) and Electrospray Ionization (ESI) quadrupole-time-of-flight Tandem Mass Spectrometry (MS/MS) for direct search of raw data using MASCOT22. Since the searches retrieved no significant match, de novo sequencing was performed and the resulting sequences were BLASTed against the non-redundant sequences, allowing SMD identification for all analyzed spots22. Only in 2009 was a proteomic study described that targeted the total protein content of the Loxosceles venom23. Although the L. intermedia venom was analyzed using Multi-Dimensional Protein Identification Technology (MudPIT)24, only 39 proteins were identified. Of these proteins, only 14 were described as toxins generally found in animal venoms23. Thus, this proteomic study seems to have severely underestimated the great toxin diversity of the Loxosceles venom, particularly in comparison to the many publications that already described distinct molecular clones from venoms of different Loxosceles species25–32. Transcriptome analyses of the L. laeta and L. intermedia venoms revealed a huge complexity of brown-spider venoms19,20. Specifically, the analysis of the L. intermedia venom described three classes of toxins comprising most toxin-encoding transcripts, such as peptides of low molecular mass (55.9%), astacin-like proteases (22.6%), and PLDs (20.2%). Also, transcripts similar to hyaluronidases, serine proteases, serine protease inhibitors, venom allergens, and members of the translationally controlled tumor protein (TCTP) family presented low levels of expression20. Although considerable information is now available on venom gland transcripts of L. intermedia, the total protein content of this venom has remained unclear. A previous study from our group applied two-dimensional immunoblots and zymograms on the venom of L. intermedia, L. laeta, and L. gaucho, and revealed several spots with differential volume containing proteins having gelatinolytic activity corresponding to astacin-like proteases33. These results corroborate that venoms from these species present a broad astacin-like family with many isoforms22,33,34.
The lack of genomic data from this arachnid prevents employing the PSM approach in full, so most of the weightlifting must be accomplished through de novo sequencing. Mainstream de novo sequencing, however, cannot efficiently handle unanticipated post-translational modifications, being far more prone to generating sequencing errors. This is because various molecules fail to provide enough mass spectral peaks during fragmentation to enable the sequencing of full peptides. To overcome these limitations, our dataset was acquired with multiple dissociation strategies applied to the same precursor (e.g., collision-induced dissociation (CID), higher-energy collisional dissociation (HCD), and electron-transfer dissociation (ETD)), thereby enabling the use of state-of-the-art de novo sequencing algorithms. These capitalize on complementary dissociation information and thus achieve unprecedented sequencing accuracy35,36. The use of different proteolytic enzymes on the venom aliquots unlocks the application of another very powerful paradigm, that of spectral networks37,38. These ‘specnets’ align spectra against one another, ultimately allowing the detection of unanticipated post-translational modifications. Moreover, they can assemble consensus mass spectra from overlapping peptides yielded by different proteolytic digests. A consensus spectrum thus obtained presents a better signal-to-noise ratio and allows for the de novo sequencing of amino-acid stretches far longer than those handled by the conventional approach. Once high-confidence de novo data are available, it becomes possible to employ tools, such as PepExplorer39 or Meta-SPS37, that apply pattern recognition approaches to the mapping of de novo sequencing data against sequences from homologous organisms, thereby facilitating biological interpretation.
By themselves, the meta-contig assemblies provided by spectral networks are not enough for one to conclude whether a biomolecule obtained 100% coverage. To pave the way in this direction, top-down proteomic data in combination with MS3 (i.e., product ion(s) selected from an MS/MS spectrum further fragmented and producing another tandem mass spectrum) and ETD were also acquired for a partition of the venom molecules into two sets (<~10 kDa and >~10 kDa). The top-down strategy consists of injecting intact proteins into the mass spectrometer, thus doing away with the inference limitations of the peptide-centric approach40. This provides complementary information to that of the networks and helps in the discovery of how much is required for obtaining full coverage. We anticipate that these data will be fundamental in the development of next-generation algorithms capable of bridging the gap between bottom-up, middle-down, and top-down proteomics.
Here, we present the first multi-protease, multi-dissociation, bottom-up-to-top-down proteomic dataset of the venom of L. intermedia, the ‘urban’ spider species commonly found in the city of Curitiba, Brazil41, along with an analysis using state-of-the-art tools. The approach stems from the motivation that multiple enzyme digestion increases protein coverage42, besides relying on different activation and acquisition methods.
Methods
Sample preparation
Adult L. intermedia specimens (both male and female) were collected in the wild in accordance with the Brazilian Federal System for Authorization and Information on Biodiversity (SISBIO-ICMBIO, license number 29801-1). Venom from 200 spiders was extracted through the electrostimulation method43 and immediately diluted in ammonium bicarbonate buffer 0.4 M/urea 8 M. Protein concentration was determined through the Coomassie blue method, using bovine serum albumin (BSA) as standard curve44. First, the venom was separated into two fractions using an ultra-filter unit (MW cutoff 10 kDa) (Millipore), one fraction containing venom proteins above ~10 kDa (400 μg) and the other containing venom proteins and peptides bellow ~10 kDa (90 μg). All procedures described next were performed equally for each fraction, after further dividing it into four aliquots, each of which was reduced with dithiothreitol (DTT) to a final concentration of 25 mM for 3 h at room temperature. Afterwards, the samples were alkylated with iodacetamide (IAA) to a final concentration of 80 mM for 15 min at room temperature in the dark. Each aliquot was digested with one of the follow enzymes: trypsin (Trypsin Gold, Mass Spectrometry Grade, Promega Corporation, Madison, cat. No. V5280, WI, USA), chymotrypsin (Promega, cat. No. V1062), and pepsin (Promega, cat. No. V1959) at the ratio of 1:50 (E:S). We note that an additional aliquot was stored and not digested. Three aliquots were incubated individually with each enzyme for 18 h, at 25 °C for chymotrypsin and 37 °C for trypsin and pepsin. The other aliquot was incubated for only 4 h with trypsin at 37 °C. Each digested fraction was divided into three aliquots and desalted with ultra-micro C-18 spin columns according to the manufacturer’s instructions (Harvard Apparatus). One of these three aliquots was stored for future use, another had its peptides desalted and directly submitted to reverse phase chromatography coupled online with an Orbitrap XL mass spectrometer. The third aliquot of the desalted peptides was eluted with 70% acetonitrile (ACN) and 0.1% formic acid, then dried in a speed vacuum concentrator, suspending buffer C (i.e., 10 mM of K2HPO4, 25%ACN, pH=3.0). Afterwards, the sample was passed through a micro strong cation exchanged spin column (SCX) according to the manufacturer’s instructions (Harvard Apparatus). Briefly, the column was equilibrated with buffer C, centrifuged for 1 min at 100×g, and the sample was eluted from the SCX spin column with increasing concentration of KCl, i.e., 100, 170, 290, and 400 mM. Finally, each fraction was desalted once more with ultra-micro C-18 spin columns according to the manufacturer’s instructions (Harvard Apparatus). All columns were then washed ten times with 0.1% formic acid and the peptides were eluted with buffer B (i.e., 70% acetonitrile, 0.1% formic acid) to proceed to next step.
Mass spectrometry analysis
Each fraction of peptides, including the non-fractionated as well as those from the SCX fractionation, was previously desalted and subjected to an LC-MS/MS analysis on a nano-LC 1D plus System (Eksigent, Dublin, CA), an ultra-high performance liquid chromatography (UHPLC) system coupled with an LTQ-Orbitrap XL ETD (Thermo, San Jose, CA) mass spectrometer, at the Mass Spectrometry Facility RPT02H of the Carlos Chagas Institute (Fiocruz, Brazil). In these analyses, the peptide mixtures were loaded onto a column (75 mm i.d., 15 cm long), packed in-house with a 3.2 μm ReproSil-Pur C18-AQ resin (Dr Maisch) with a flow of 500 nl/min and subsequently eluted with a flow of 250 nl/min from 5 to 40% ACN in 0.5% formic acid in a 120 min gradient. The mass spectrometer was set to data-dependent mode to automatically switch between MS and MS/MS acquisition. Full-scan MS spectra (m/z 350–1,800) were acquired in the Orbitrap analyzer with resolution R=60,000 at m/z 400 (after accumulation to a target value of 1,000,000 in the linear trap) using survey mode. The three most intense ions were sequentially isolated and fragmented using CID, HCD, and ETD for the same precursor. Previous target ions selected for MS/MS were dynamically excluded for 60 s. The total cycle time was approximately 5 s. The general mass spectrometric conditions were: spray voltage, 2.4 kV; no sheath or auxiliary gas flow; ion transfer tube temperature, 100 °C; collision gas pressure, 1.3 mTorr; normalized collision energy using wide-band activation mode; 35% for MS/MS. Ion selection thresholds were of 5,000 counts for MS/MS. The parameters for each fragmentation type in MS/MS acquisitions were as follows. For CID: isolation width, m/z 2.5; normalized collision energy, 35; activation, q=0.25; activation time, 30 ms. For HCD: isolation width, m/z 2.5; normalized collision energy, 35; activation time, 30 ms; full width at half maximum resolution, 15,000. For ETD: isolation width, m/z 2.5; activation time, 100 ms.
Bioinformatics analysis
The de novo sequencing approach employed in this work utilized multiple MS/MS spectra from overlapping peptides, generated from multiple proteases and of precursors analyzed with CID, HCD, and ETD spectrum triples. Each was then converted into prefix residue mass (PRM) spectra. In this conversion, MS/MS peak masses were converted into putative cumulative precursor fragment masses, with intensity scores determined from likelihood models specific to each fragmentation mode. Triples of PRM spectra from the same precursor were then merged into a single PRM spectrum per precursor by adding scores for matching peak masses. Spectral-network algorithms, implemented in the ProteoSAFe web platform that is freely accessible at http://proteomics.ucsd.edu/ProteoSAFe/, were then used to align merged PRM spectra from peptides with overlapping sequences. Moreover, A-Bruijn algorithms were used to integrate these alignments into assembled contigs.
Each contig was then used to construct a consensus contig spectrum, or meta-contig, capitalizing on the corroborating evidence from all of its assembled spectra to yield a high-quality consensus de novo sequence36. Subsequently, the Meta-SPS algorithm was used to align the meta-contigs against a FASTA sequence database37. This database contained all Loxosceles sequences from UniProt, all from the transcriptome of the L. intermedia venom gland20, and an internal database with common mass spectrometry contaminants and proteases.
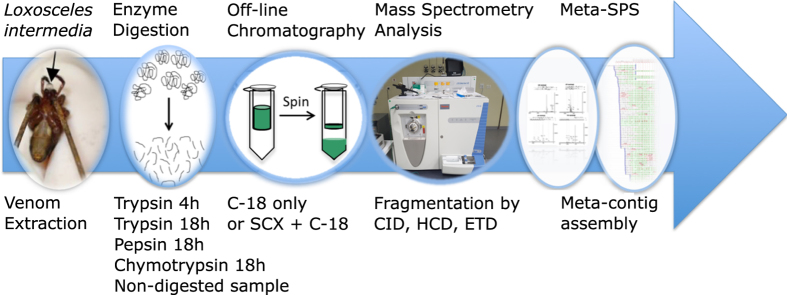
A summary of this methodology is found in Fig. 1.
Figure 1. Methodology workflow.
Summary of the sequence of procedures that constitute the methodology employed, from venom extraction to the meta-contig assembling that enabled the identifications of venom proteins.
Data Records
Our bioinformatics analysis disclosed a list of 190 proteins (Table 1). As far as we know, this is the most complete comprehensive proteomic profiling of the L. intermedia venom. All mass spectrometry data are available from both the ProteomeXchange Consortium via the PRIDE45 partner repository, with dataset identifier PXD005523 (Data Citation 1), and our servers (http://proteomics.fiocruz.br/pcarvalho/lintermedia/venom/). A full list of the proteins, meta-contigs, and homologous sequences is made available in Table 1.
Table 1. Identifications resulting from the >~10 kDa and <~10 kDa venom fractions.
| >~10 kDa | <~10 kDa | Together | |
|---|---|---|---|
| The last column, Together, eliminates redundancies (viz., by maximum parsimony53), as well as contaminants and the proteases used during sample preparation (trypsin, chymotrypsin, and pepsin). | |||
| No. of spectral triplets | 41,386 | 36,625 | 78,011 |
| No. of contigs | 642 | 454 | 1,096 |
| No. of homologous sequences | 440 | 228 | 190 |
| No. of de novo sequences | 642 | 454 | 1,096 |
| No. of proteins | 440 | 228 | 190 |
| No. of raw files | 42 | 46 | 88 |
All Meta-SPS results for >~10 kDa and <~10 kDa, together with the parameter files used for running the software, are available as separate material (MetaSPS_Results.xlsx, Data Citation 2). The results are presented in six tabs, viz., for >~10 kDa grouped by contig, >~10 kDa grouped by spectrum, >~10 kDa parameter file, <~10 kDa grouped by contig, <~10 kDa grouped by spectrum, and <~10 kDa parameter file.
Technical Validation
The lack of any previous comprehensive proteomic analysis of the Loxosceles venom demonstrates that studying this venom in detail has been a challenge, one that stems from the organism being highly non-canonical and from the fact that protein sequences for it have remained scarce in databases. The present work circumvented these obstacles by using a combination of shotgun proteomic experiments and different tools to generate and analyze large proteomic datasets and de novo sequencing results.
Our results revealed 190 protein identifications, including all classes of toxins described in previous transcriptome analyses19,20 (Table 2 (available online only)). Our approach identified both high- and low-abundance toxins of the L. intermedia venom, as well as homolog sequences from distinct Loxosceles species (astacin-like proteases, PLDs, peptides, TCTPs, hyaluronidases, allergens, serine proteases, serine protease inhibitors, and housekeeping proteins) (Table 2 (available online only)). These data reinforce the holocrine nature of the Loxosceles venom gland23 and demonstrate that its venom is composed of toxins and housekeeping proteins originating from epithelial-cell content, such as the angiotensin converting enzyme, the 60S ribosomal protein, the Na-Pi co-transporter, and the myosin heavy chain (Table 2 (available online only)). Our results, therefore, validate the method used for analyzing the proteome of an organism with non-sequenced genome.
Table 2. Meta-contig assemblies output by Meta-SPS on shotgun data from the venom of Loxosceles intermedia and corresponding proteins mapped per similarity.
| de novo sequence | Protein index | Protein name | Species | |
|---|---|---|---|---|
| The contigs are listed according to their toxin class. | ||||
| Toxin class |
||||
| Metalloprotease | [211.812][154.295][261.952]Y[200.869]NMIYGAG | gi 116733934 gb ABK20019.1 | astacin-like metalloprotease toxin precursor | Loxosceles intermedia |
| [168.09]PDATIVY[128.106] | gi 257219865 gb ACV52010.1 | astacin-like metalloprotease toxin 2 precursor | Loxosceles intermedia | |
| DDYVI[262.263][128.148] | gi 257219867 gb ACV52011.1 | astacin-like metalloprotease toxin 3 precursor | Loxosceles intermedia | |
| GVIIHE[291.835][227.31]GFY | TSA 1, Contig: LIS109 | putative astacin-like metalloprotease | Loxosceles intermedia | |
| GTCICDGGSCNC[128.125]R | TSA 1, Contig: LIS137 | putative astacin-like metalloprotease | Loxosceles intermedia | |
| [127.832]VRNGCNT[252.829]DVGIR | TSA 1, Contig: LIS187 | putative astacin-like metalloprotease | Loxosceles intermedia | |
| RDAP[215.036][170.707][196.097]IG[153.721]F | TSA 2, Contig: LIC247 | putative astacin-like metalloprotease | Loxosceles intermedia | |
| GNMFSCWSTVGMR | TSA 2, Contig: LIC331 | putative astacin-like metalloprotease | Loxosceles intermedia | |
| INIIY[128.088] | TSA 2, Contig: LIS63 | putative astacin-like metalloprotease | Loxosceles intermedia | |
| [128.115]PCGNNV[274.883]DP[227.119] | TSA 3, Contig: LIC321 | putative astacin-like metalloprotease | Loxosceles intermedia | |
| GAAGVTIINPAAGR | TSA 3, Contig: LIC369 | putative astacin-like metalloprotease | Loxosceles intermedia | |
| NAVSSD[244.341] | TSA 3, Contig: LIS121 | putative astacin-like metalloprotease | Loxosceles intermedia | |
| GVIIHEIGHVIGF | sp A0FKN6 VMPA LOXIN | astacin-like metalloprotease toxin precursor | Loxosceles intermedia | |
| [153.983]IDI[128.103][241.226]NTIY | tr C9D7R2 C9D7R2 LOXIN | astacin-like metalloprotease toxin 2 precursor | Loxosceles intermedia | |
| GDSRIWPDGVVIY | tr C9D7R3 C9D7R3 LOXIN | astacin-like metalloprotease toxin 3 precursor | Loxosceles intermedia | |
| F[242.232]D[275.039][128.008]RCVY | TSA 1, Contig: LIC313 | putative astacin-like metalloprotease | Loxosceles intermedia | |
| [127.931]D[128.061][241.133]DR[184.194][174.03]PY | TSA 1, Contig: LIC411 | putative astacin-like metalloprotease | Loxosceles intermedia | |
| IEAGDV[128.029]GIGCGG | TSA 1, Contig: LIS117 | putative astacin-like metalloprotease | Loxosceles intermedia | |
| [349.495]IGITGF | TSA 1, Contig: LIS234 | putative astacin-like metalloprotease | Loxosceles intermedia | |
| [184.051]N[257.098][216.857]I[259.978]E[291.043]SIPSN | TSA 1, Contig: LIS98. | putative astacin-like metalloprotease | Loxosceles intermedia | |
| F[127.959]NHDS[204.387]D[242.74] | TSA 2, Contig: LIC228 | putative astacin-like metalloprotease | Loxosceles intermedia | |
| RDAP[215.036][170.707][196.097]IG[153.721]F | TSA 2, Contig: LIC247 | putative astacin-like metalloprotease | Loxosceles intermedia | |
| GIGIT[128.015]PCTCS[198.07]C[225.217] | TSA 2, Contig: LIC331 | putative astacin-like metalloprotease | Loxosceles intermedia | |
| INIIY[128.088] | TSA 2, Contig: LIS63 | putative astacin-like metalloprotease | Loxosceles intermedia | |
| Phospholipase-D | [198.061]YITAST[234.471]D[127.861][128.324]DFA[128.058] | gi 141452623 gb ABO87656.1 | dermonecrotic toxin isoform 6 | Loxosceles intermedia |
| HYEIF[128.043]GFR | gi 156067386 gb ABU43333.1 | loxtox i5 | Loxosceles intermedia | |
| [241.121]ICAIVI[202.255]G[245.305]II[326.299] | gi 156067390 gb ABU43335.1 | loxtox i7 | Loxosceles intermedia | |
| [185.016]CC[215.047]DVANAEAW | gi 224472025 gb ACN48894.1 | sphingomyelinase D-like protein, partial | Loxosceles arizonica | |
| VATYDDN[283.158]V[248.057][128.175][128.114] | gi 224472109 gb ACN48936.1 | sphingomyelinase D-like protein, partial | Loxosceles spadicea | |
| I[128.164][185.033][320.151][199.846]V[128.057]V[257.079] | gi 224472111 gb ACN48937.1 | sphingomyelinase D-like protein, partial | Loxosceles variegata | |
| [258.156]GS[215.905]C[128.131]TN[142.105] | gi 224472117 gb ACN48940.1 | sphingomyelinase D-like protein, partial | Loxosceles arizonica | |
| [323.223]SIDIIAS[128.107] | gi 224472133 gb ACN48948.1 | sphingomyelinase D-like protein, partial | Loxosceles hirsuta | |
| WVI[211.923][323.19][193.88]G[184.835]DWG[288.885]AGVVGGIV[168.053] | gi 224472141 gb ACN48952.1 | sphingomyelinase D-like protein, partial | Loxosceles rufescens | |
| IA[277.273]D[217.017]F[286.991]V[275.199] | gi 224472143 gb ACN48953.1 | sphingomyelinase D-like protein, partial | Loxosceles arizonica | |
| [127.947]IAEWFDVDVC | gi 224472147 gb ACN48955.1 | sphingomyelinase D-like protein, partial | Loxosceles laeta | |
| I[172.141]NFMN[128.005]R | gi 224472157 gb ACN48960.1 | sphingomyelinase D-like protein, partial | Loxosceles laeta | |
| AGADGM[212.996]DFP[127.998] | gi 224472195 gb ACN48979.1 | sphingomyelinase D-like protein, partial | Loxosceles aff. Spinulosa GJB-2008 | |
| [128.05][346.235]FGWEIC[128.103] | gi 224472201 gb ACN48982.1 | sphingomyelinase D-like protein, partial | Loxosceles aff. Spinulosa GJB-2008 | |
| I[128.005]NYWNNGDNG[230.782] | gi 225008387 gb ACN48920.2 | sphingomyelinase D-like protein, partial | Loxosceles sabina | |
| [234.124]IIISI[211.97]T[220.159]Y[128.131] | gi 225008389 gb ACN48924.2 | sphingomyelinase D-like protein, partial | Loxosceles variegata | |
| [217.067]VDDGS[127.933][261.153]IGGDSCC[127.973] | gi 225008391 gb ACN48959.2 | sphingomyelinase D-like protein, partial | Loxosceles laeta | |
| [202.038]TYEDN[283.238]VTF[128.109]A | gi 49458048 gb AAT66074.1 | sphingomyelinase D-like protein 3, partial | Loxosceles boneti | |
| WENF[229.222]FI[128.141] | gi 49458050 gb AAT66075.1 | sphingomyelinase D protein 1, partial | Loxosceles reclusa | |
| IATYDDN[283.149][128.13][128.098] | gi 49458052 gb AAT66076.1 | sphingomyelinase D protein 2, partial | Loxosceles reclusa | |
| WSR[225.203]IWDIAHM | gi 81343346 gb ABB71184.1 | dermonecrotic toxin isoform 3 | Loxosceles intermedia | |
| [128.059]SSI[185.218]DN[127.961]AY[128.189]AGVNMATDI[275.181] | gi 90192366 gb ABD91846.1 | dermonecrotic toxin isoform 4 | Loxosceles intermedia | |
| [259.057]SFADYIDYMR | gi 90192368 gb ABD91847.1 | dermonecrotic toxin isoform 5 | Loxosceles intermedia | |
| AIC[262.118]N[213.308][157.942]IF[269.463]M | TSA 1, Contig: LIC336 | putative phospholipase-D | Loxosceles intermedia | |
| [128.065]PI[257.302][168.016][128.103] | TSA 1, Contig: LIS80 | putative phospholipase-D | Loxosceles intermedia | |
| WVI[213.173]FE[230.5]VEDWG[289.028]AGN[198.021]IV[168.053] | TSA 3, Contig: LIC182 | putative phospholipase-D | Loxosceles intermedia | |
| CENISTDD[214.057]R | TSA 3, Contig: LIC203 | putative phospholipase-D | Loxosceles intermedia | |
| R[169.813][231.411]ANPIGR | TSA 3, Contig: LIC334 | putative phospholipase-D | Loxosceles intermedia | |
| [127.988]AE[300.164]D[128.052]IF[128.126]I[255.151] | TSA 3, Contig: LIC352 | putative phospholipase-D | Loxosceles intermedia | |
| WVIGG[184.011]A[216.684][200.309]T[200.21][184.253][200.309][216.684]GVVGGIV[168.237] | TSA 3, Contig: LIC395 | putative phospholipase-D | Loxosceles intermedia | |
| [255.268]TCEYIEI[245.969]DSNYSEIG[224.144] | TSA 3, Contig: LIC419 | putative phospholipase-D | Loxosceles intermedia | |
| GEEYVNVFPMGIR | TSA 3, Contig: LIC423 | putative phospholipase-D | Loxosceles intermedia | |
| [256.067][269.811][259.263][227.115]GHEPHC[299.732] | TSA 3, Contig: LIS9 | putative phospholipase-D | Loxosceles intermedia | |
| [128.078][128.124]AGV[128.053]D[128.036]EHIW | sp A4USB4 A51 LOXIN | phospholipase D LiSicTox-alphaV1 Dermonecrotic toxin 6 | Loxosceles intermedia | |
| Toxin class |
||||
| Phospholipase-D | [278.133]A[127.914]ARDAG[128.044]V[127.924] | sp B2KKW0 A22 LOXIN | phospholipase D LiSicTox-alphaII2 Loxtox i5 | Loxosceles intermedia |
| TARDVA | sp C0JAQ5 A1IA1 LOXHI | phospholipase D LhSicTox-alphaIA2ai Dermonecrotic toxin | Loxosceles hirsuta | |
| [144.086][196.168][128.016][299.803][217.693]DNGNN[142.216][128.015] | sp C0JAR3 A1IA6 LOXHI | phospholipase D LhSicTox-alphaIA2avi Dermonecrotic toxin | Loxosceles hirsuta | |
| PI[128.15]TD[276.095][260.159][226.968] | sp C0JAR7 A1IA7 LOXHI | phospholipase D LhSicTox-alphaIA2avii Dermonecrotic toxin | Loxosceles hirsuta | |
| [128.069][314.204][266.04]EIIE[128.136]VGY | sp C0JAS6 A1I1 LOXSP | phospholipase D LspaSicTox-alphaIA2i Dermonecrotic toxin | Loxosceles spadicea | |
| [128.135]G[238.403]YEDNPW | sp C0JAT4 A1H1 LOXHI | phospholipase D LhSicTox-alphaIA1i Dermonecrotic toxin | Loxosceles hirsuta | |
| [285.246]TIT[127.957][128.38][193.725]PE[244.902][128.249]F[300.312] | sp C0JAU6 A1LC LOXAR | phospholipase D LarSicTox-alphaIB2c Dermonecrotic toxin | Loxosceles arizonica | |
| IISI[211.095]ID[300.088] | sp C0JAV3 A1KA1 LOXAP | phospholipase D LapSicTox-alphaIB1ai Dermonecrotic toxin | Loxosceles apachea | |
| [128.046]T[261.162][127.9]SAG | sp C0JAX3 A1MA1 LOXDE | phospholipase D LdSicTox-alphaIB3ai Dermonecrotic toxin | Loxosceles deserta | |
| YWTVD[128.227]Y | sp C0JAY0 A1MA6 LOXDE | phospholipase D LdSicTox-alphaIB3ai Dermonecrotic toxin | Loxosceles deserta | |
| GIIISI[250.013]IAHY | sp C0JAZ1 A1OA1 LOXVA | phospholipase D LvSicTox-alphaIC1ai Dermonecrotic toxin | Loxosceles variegata | |
| NAIETDVT | sp C0JAZ4 A1OB1 LOXVA | phospholipase D LvSicTox-alphaIC1bi Dermonecrotic toxin | Loxosceles variegata | |
| [127.882]GI[185.036]EGC[200.002][372.207]ICA | sp C0JAZ8 A1OB5 LOXVA | phospholipase D LvSicTox-alphaIC1bv Dermonecrotic toxin | Loxosceles variegata | |
| R[227.227]TT[154.041]V | sp C0JB02 A1OD LOXRU | phospholipase D LruSicTox-alphaIC1d Dermonecrotic toxin | Loxosceles rufescens | |
| GEND[170.749]N[270.185]AY | sp C0JB04 A1P LOXRU | phospholipase D LruSicTox-alphaIC2 Dermonecrotic toxin | Loxosceles rufescens | |
| [128.095]EVIGVTII[143.89]TCEAH[252.229][210.281]D[274.244] | sp C0JB05 A21 LOXSP | phospholipase D LspaSicTox-alphaII1 Dermonecrotic toxin | Loxosceles spadicea | |
| FCGC[128.12]AWNPGHC[261.261][127.94] | sp C0JB06 A21 LOXVA | phospholipase D LvSicTox-alphaII1 Dermonecrotic toxin | Loxosceles variegata | |
| Y[293.065]PCDCF | sp C0JB07 A21 LOXAP | phospholipase D LapSicTox-alphaII1 Dermonecrotic toxin | Loxosceles apachea | |
| R[312.174][247.035][256.156]AVN | sp C0JB09 A31 LOXAR | phospholipase D LarSicTox-alphaIII1 Dermonecrotic toxin | Loxosceles arizonica | |
| [349.464]NDGCP[314.14]CNDW | sp C0JB12 A332 LOXLA | phospholipase D LlSicTox-alphaIII3ii Dermonecrotic toxin | Loxosceles laeta | |
| N[128.122]AGV[128.154]DREHVW | sp C0JB14 A411 LOXHI | phospholipase D LhSicTox-alphaIV1i Dermonecrotic toxin | Loxosceles hirsuta | |
| VGGSCNDD[127.823]VCC[128.169]GG[128.002] | sp C0JB18 A411 LOXAM | phospholipase D LamSicTox-alphaIV1i Dermonecrotic toxin | Loxosceles amazonica | |
| RIANYDD[345.981][289.109]F | sp C0JB22 A41 LOXAR | phospholipase D LarSicTox-alphaIV1 Dermonecrotic toxin | Loxosceles arizonica | |
| WFDVDVC[128.184]GG | sp C0JB23 A411 LOXLA | phospholipase D LlSicTox-alphaIV1i Dermonecrotic toxin | Loxosceles laeta | |
| [256.225]F[315.247]N[251.495]W[128.071][171.981]GI[214.973] | sp C0JB25 A421 LOXLA | phospholipase D LlSicTox-alphaIV2i Dermonecrotic toxin | Loxosceles laeta | |
| [201.075]DDD[183.982]D[342.088][248.103][282.879]W[260.214] | sp C0JB29 A43 LOXLA | phospholipase D LlSicTox-alphaIV3 Dermonecrotic toxin | Loxosceles laeta | |
| FI[128.05]GDYINV | sp C0JB30 A71 LOXAR | phospholipase D LarSicTox-alphaVII1 Dermonecrotic toxin | Loxosceles arizonica | |
| [127.979]SY[168.058]VIVG[326.398]E[257.168][128.109]D[291.254] | sp C0JB31 A611 LOXHI | phospholipase D LhSicTox-alphaVI1i Dermonecrotic toxin | Loxosceles hirsuta | |
| [243.149]R[214.248]D[228.235]I[128.113][128.353][226.9]TIY | sp C0JB40 B1O LOXCS | phospholipase D LcsSicTox-betaIC1 Dermonecrotic toxin | Loxosceles cf. spinulosa GJB-2008 | |
| [249.996]S[320.07]C[292.123]PMI[303.312]V | sp C0JB44 B1T1 LOXSN | phospholipase D LspiSicTox-betaIE3i Dermonecrotic toxin | Loxosceles spinulosa | |
| [260.294]C[274.817]N[250.295]N[229.03][250.931]INNR[248.338] | sp C0JB48 B1S LOXAS | phospholipase D LafSicTox-betaIE2 Dermonecrotic toxin | Loxosceles cf. spinulosa GJB-2008 | |
| [128.075]GP[260.085][128.08]FNPGNYDEEE[269.243] | sp C0JB92 B31 LOXSN | phospholipase D LspiSicTox-betaIII1 Dermonecrotic toxin | Loxosceles spinulosa | |
| HGIPCDCGRSCI | sp P0CE78 A1H1 LOXRE | phospholipase D LrSicTox-alphaIA1i Dermonecrotic toxin | Loxosceles reclusa | |
| [185.577]ERR[210.449]WIMG[220.232] | sp P0CE79 A1H2 LOXRE | phospholipase D LrSicTox-alphaIA1ii Dermonecrotic toxin | Loxosceles reclusa | |
| ADNRG[196.202]IW | sp P0CE80 A1HA LOXIN | phospholipase D LiSicTox-alphaIA1a Dermonecrotic toxin 1 | Loxosceles intermedia | |
| E[310.079]DRV[128.206][127.978] | sp P0CE83 A1IA1 LOXIN | phospholipase D LiSicTox-alphaIA2ai Dermonecrotic toxin LiP2 | Loxosceles intermedia | |
| RN[172.129]A[185.125]IIMAVI | sp Q1KY79 A32 LOXLA | phospholipase D LlSicTox-alphaIII2 Dermonecrotic toxin Ll2 | Loxosceles laeta | |
| [128.089]SRD[226.149]DHIW | sp Q1W694 B1Q LOXIN | phospholipase D LiSicTox-betaID1 Dermonecrotic toxin 5 | Loxosceles intermedia | |
| [241.168]D[128.065][246.052]D[306.802][314.355]EF | sp Q1W695 A21 LOXIN | phospholipase D LiSicTox-alphaII1 Dermonecrotic toxin 4 | Loxosceles intermedia | |
| [144.192][184.287][171.423][244.328][212.328]AFTDD[157.983][314.318] | sp Q27Q54 B1H2 LOXIN | phospholipase D LiSicTox-betaIA1ii Dermonecrotic toxin-like II | Loxosceles intermedia | |
| [271.172]AI[141.933][167.905][170.252]EEI[276.104][127.878] | sp Q2XQ09 B1H1 LOXIN | phospholipase D LiSicTox-betaIA1i Dermonecrotic toxin-like I | Loxosceles intermedia | |
| Y[128.246]E[128.214]IIIF | sp Q5YD77 A1KA LOXBO | phospholipase D LbSicTox-alphaIB1a Dermonecrotic toxin Lb1 | Loxosceles boneti | |
| M[169.966]D[227.167]DIA[204.21][238.193]N | sp Q8I912 B1H LOXLA | phospholipase D LlSicTox-betaIA1 Dermonecrotic toxin LlH10 | Loxosceles laeta | |
| [252.158][172.24]AG[128.07]IIS[346.153] | sp Q8I913 A331 LOXLA | phospholipase D LlSicTox-alphaIII3i Dermonecrotic toxin LlH13 | Loxosceles laeta | |
| I[248.195][172.042]RTNCC[317.124][316.196] | tr G8GZ61 G8GZ61 9ARAC | Sphingomyelinase D A1 | Loxosceles adelaida | |
| GMDIPNIRI[198.131][274.115] | TSA 2, Contig: LIS181 | putative phospholipase-D | Loxosceles intermedia | |
| IADYE[245.445]RGF | TSA 3, Contig: LIC316 | putative phospholipase-D | Loxosceles intermedia | |
| [229.079]VNDYDCA[278.03][197.89][257.788]N[341.945]F[188.387]GGI[173.846]E[203.936][227.312][212.04] | TSA 1, Contig: LIS124 | putative phospholipase-D | Loxosceles intermedia | |
| ICK-peptide | [288.103][276.948]A[228.135]PETA[217.192]E[258.063]GH[270.214] | gi 118574181 sp Q27Q53.1 TX4 LOXIN | U1-sicaritoxin-Li1c LiTx4 | Loxosceles intermedia |
| [141.867]GCTMGVC[262.034]G[127.99] | gi 74786589 sp Q6B4T3.1 TX3 LOXIN | U2-sicaritoxin-Li1a LiTx3 | Loxosceles intermedia | |
| [170.014]T[257.226][128.082]CPAWSHER | gi 74786590 sp Q6B4T4.1 TX2 LOXIN | U1-sicaritoxin-Li1b LiTx2 | Loxosceles intermedia | |
| [128.076][198.175]C[128.129][257.053]AWSH[285.151]ECR | gi 74786591 sp Q6B4T5.1 TX1 LOXIN | U1-sicaritoxin-Li1a LiTx1 | Loxosceles intermedia | |
| [242.133]VECICSPSYYP[154.122][128.073] | TSA 1, Contig: LIC275 | putative LiTx3 | Loxosceles intermedia | |
| [128.047]GI[220.146][299.366]THH[128.119]Y[196.17]E[316.185] | TSA 1, Contig: LIC298 | putative LiTx3 | Loxosceles intermedia | |
| WVI[213.183]FE[230.39][229.002]DWG[288.987][127.905]VV[128.365]V[285.053] | TSA 1, Contig: LIS111 | putative LiTx3 | Loxosceles intermedia | |
| [174.128]TVPVYAECGR | TSA 1, Contig: LIS14 | putative LiTx3 | Loxosceles intermedia | |
| [128.108][128.122]NVMRIYVG | TSA 1, Contig: LIS64 | putative LiTx3 | Loxosceles intermedia | |
| AGGASE[288.32]V[288.068]E | TSA 1, Contig: LIC255 | putative LiTx4 | Loxosceles intermedia | |
| Toxin class |
||||
| ICK-peptide | [174.137]GIPNASGSIGR | TSA 2, Contig: LIC315 | putative LiTx3 | Loxosceles intermedia |
| R[217.187][200.251]FVPVG[174.181]G | TSA 2, Contig: LIS1 | putative LiTx3 | Loxosceles intermedia | |
| GAD[225.773][283.154][216.215] | TSA 2, Contig: LIS161 | putative LiTx3 | Loxosceles intermedia | |
| [342.104][142.018][174.19][275.359]TCHGPNWAA[342.104] | TSA 2, Contig: LIS33 | putative LiTx3 | Loxosceles intermedia | |
| [371.939]WNYA[199.308][128.365]STII | TSA 3, Contig: LIC212 | putative LiTx3 | Loxosceles intermedia | |
| [128.037]SSFEDF[227.173]VDCNS[261.16] | TSA 3, Contig: LIS21 | putative LiTx3 | Loxosceles intermedia | |
| G[216.293]CSDG[342.128]DIPC[128.288] | TSA 3, Contig: LIS35 | putative LiTx3 | Loxosceles intermedia | |
| IGISSD[320.03]PDW[128.125] | TSA 3, Contig: LIS36 | putative LiTx3 | Loxosceles intermedia | |
| ECI[241.167][241.217][345.95] | TSA 3, Contig: LIS43 | putative LiTx3 | Loxosceles intermedia | |
| AAGDTN[259.089][229.905][237.813][320.42]W[128.09] | TSA 3, Contig: LIS44 | putative LiTx3 | Loxosceles intermedia | |
| RCE[224.918]GI[128.1]DISE | TSA 3, Contig: LIS57 | putative LiTx3 | Loxosceles intermedia | |
| [226.296]PNI[128.014][185.165]T[247.019]CNN[226.997] | TSA 3, Contig: LIC381 | putative neurotoxin like-magi-3 | Loxosceles intermedia | |
| [258.986]VCYC[208.258]FGV[128.123]NC[128.024] | TSA 3, Contig: LIS20 | putative neurotoxin like-magi-3 | Loxosceles intermedia | |
| GIRSATTPGNA[128.185]Y | sp P0CE83 A1IA1 LOXIN | phospholipase D LiSicTox-alphaIA2ai Dermonecrotic toxin LiP2 | Loxosceles intermedia | |
| [199.081]YD[225.268]DDWCCG[199.016] | sp Q27Q53 TX4 LOXIN | U1-sicaritoxin-Li1c LiTx4 | Loxosceles intermedia | |
| [213.19][128.003]Y[127.926][299.318][275.069]R[244.958][183.934][277.077][128.226]G[141.885] | sp Q6B4T3 TX3 LOXIN | U2-sicaritoxin-Li1a LiTx3 | Loxosceles intermedia | |
| CT[224.823]CGPYY | sp Q6B4T4 TX2 LOXIN | U1-sicaritoxin-Li1b LiTx2 | Loxosceles intermedia | |
| [320.096][128.127][128.065]GTPC[128.107]CPAWSHER[289.113][174.07] | sp Q6B4T5 TX1 LOXIN | U1-sicaritoxin-Li1a LiTx1 | Loxosceles intermedia | |
| TG[128.074][128.206]FI | TSA 1, Contig: LIS15 | putative LiTx1 | Loxosceles intermedia | |
| [260.124][257.99][260.764]DAIESEDPV[208.163] | TSA 1, Contig: LIS159 | putative LiTx1 | Loxosceles intermedia | |
| SV[127.961][200.118]GI[315.972][128.206]E[142.111] | TSA 1, Contig: LIS163 | putative LiTx3 | Loxosceles intermedia | |
| M[247.954][271.519]E[198.253][128.081]D[200.22] | TSA 2, Contig: LIS31 | putative LiTx3 | Loxosceles intermedia | |
| GAG[143.963]NIFA[127.947] | TSA 2, Contig: LIS76 | putative LiTx3 | Loxosceles intermedia | |
| [127.704]T[128.06]FCV[128.02]NG[128.165]PICP[128.162][167.895]G | TSA 3, Contig: LIC337 | putative LiTx1 | Loxosceles intermedia | |
| G[266.174]CHAFGSNCR | TSA 2, Contig: LIS229 | putative neurotoxin like-magi-3 | Loxosceles intermedia | |
| Serine protease inhibitor | [128.02]CP[235.787][198.175][143.863]G[241.815]DV[127.896][218.419]G | TSA 1, Contig: LIS209 | putative serine protease inhibitor | Loxosceles intermedia |
| Serine protease | [269.335][262.236]S[244.714]WASFP[211.817]SA[128.235]I[269.335] | TSA 1, Contig: LIC305 | putative serine protease | Loxosceles intermedia |
| Venom allergen | [346.178][257.117]N[215.219][298.78][217.218]ND[260.144]IG[256.421][372.04] | TSA 2, Contig: LIC179 | putative venom allergen | Loxosceles intermedia |
| Hyaluronidase | V[207.937]SSEY[211.831]I[326.363][319.484] | TSA 1, Contig: LIS222 | putative hyaluronidase | Loxosceles intermedia |
| TCTP | [256.849][275.401][210.175]I[252.242]TA[244.108][200.309][241.062]N[170.078]IIED[372.344] | tr G3LU44 G3LU44 LOXIN | translationally-controlled tumor protein homolog LiTCTP | Loxosceles intermedia |
| [269.086]FGIMAP[225.403]GEIR | gi 344995179 gb AEN55462.1 | translationally controlled tumor protein LiTCTP | Loxosceles intermedia | |
| Cellular processes proteins | [200.059]VHEDNI[128.065]E[128.062]H[128.193][262.127] | TSA 1, Contig: LIC216 | putative 60S ribosomal protein L10 | Loxosceles intermedia |
| [199.078]CT[297.052]F[264.084][233.709]GCPSR | TSA 1, Contig: LIS275 | putative cysteine-rich PDZ-binding protein | Loxosceles intermedia | |
| [269.045]D[173.858]DPDCDC[211.944]D[173.937]D[269.044] | TSA 1 Contig: LIS239 | putative cytochrome P450 mRNA | Loxosceles intermedia | |
| I[204.06]CV[128.097][297.04] | TSA 1, Contig: LIS199 | putative histone H2B | Loxosceles intermedia | |
| [241.241][210.11]DFYEI[170.035]SA[262.036] | TSA 1, Contig: LIS189 | putative myosin heavy chain | Loxosceles intermedia | |
| [128.159]GF[218.024]D[127.978]VVIA | TSA 1, Contig: LIC349 | putative secreted salivary gland peptide | Loxosceles intermedia | |
| [262.073]I[243.142]PSNPSCR | TSA 2, Contig: LIC232 | putative 60S ribosomal protein L27 | Loxosceles intermedia | |
| [216.109]IVISNPDINH[260.307][226.167][128.074] | TSA 2, Contig: LIC283 | putative actin related protein 3 | Loxosceles intermedia | |
| TED[128.098]V[229.139] | TSA 2, Contig: LIS258 | putative glycoprotein hormone alpha-2 precurso | Loxosceles intermedia | |
| [128.037]SSFEDF[227.173]VDCNS[261.16] | TSA 2, Contig: LIS217 | putative myosin light chain | Loxosceles intermedia | |
| [199.18]SIDIIAS[128.129]DVMDR | TSA 2, Contig: LIS39 | putative Na/Pi co-transporter | Loxosceles intermedia | |
| [269.086]FGIMAP[225.403]GEIR | TSA 2, Contig: LIS280 | putative ribosomal protein L18a | Loxosceles intermedia | |
| [247.11]G[128.088]I[183.991]N[210.738][262.174][285.2] | TSA 2, Contig: LIS213 | putative ribosomal protein S3 | Loxosceles intermedia | |
| Y[127.918]GV[143.679]I[196.241]AGR | TSA 3, Contig: LIC307 | putative angiotensin-converting enzyme | Loxosceles intermedia | |
| VGSIPIDI[128.058] | TSA 3, Contig: LIC368 | putative myosin light chain | Loxosceles intermedia | |
| [128.071]SAI[250.284]IS[127.926]N | TSA 3, Contig: LIC207 | putative troponin C | Loxosceles intermedia | |
| GTIPVT[323.12]P[128.197] | TSA 3, Contig: LIS253 | putative troponin T | Loxosceles intermedia | |
| [128.057]CW[283.922]E[127.782][128.342]GA | TSA 3, Contig: LIS216 | putative zinc finger protein | Loxosceles intermedia | |
| [128.066]CG[169.924]S[293.695]D[211.062]C[143.962] | TSA 1, Contig: LIS257 | putative cAMP-responsive element-binding protein-like 2-like | Loxosceles intermedia | |
| HVSV[128.009] | TSA 1, Contig: LIC219 | putative glutamine synthetase | Loxosceles intermedia | |
| [262.165]P[230.085]D[128.062] | TSA 2, Contig: LIS220 | putative 60S ribosomal protein L6 | Loxosceles intermedia | |
| [268.215]HCTCDDVC[128.023]R[275.259]Y | TSA 2, Contig: LIS231 | putative calmodulin | Loxosceles intermedia | |
| [274.04]TPIRIN[210.228]II[228.096] | TSA 2, Contig: LIS27 | putative Na/Pi co-transporter | Loxosceles intermedia | |
| [372.255]DEIIN[171.15]IE[200.054]E[201.127]S[226.48]HG[293.174][372.288] | TSA 2, Contig: LIS265 | putative selenoprotein M | Loxosceles intermedia | |
| NW[211.918]RIR[298.994][248.176] | TSA 2, Contig: LIS205 | putative ubiquitin | Loxosceles intermedia | |
| [253.085]IG[200.301]T[251.496][171.357]ED | tr Q6W974 Q6W974 LOXRE | codium/potassium ATPase alpha subunit | Loxosceles reclusa | |
| YHNN[232.454]ISII | tr B8R316 B8R316 9ARAC | NADH-ubiquinone oxidoreductase chain 1 | Loxosceles baja | |
| [276.09]MISMEVG[307.07][259.494]Y | tr B8R320 B8R320 LOXDE | NADH-ubiquinone oxidoreductase chain 2 | Loxosceles deserta | |
| Cellular processes proteins |
||||
| [128.205]MFTIIM[235.906]GV[215.128] | tr B8R326 B8R326 9ARAC | NADH-ubiquinone oxidoreductase chain 1 | Loxosceles chinateca | |
| [245.288]RA[260.078]NTSTIGTA[212.046]R | tr B8R332 B8R332 9ARAC | NADH-ubiquinone oxidoreductase chain 1 | Loxosceles kaiba | |
| S[246.297][301.019]IVS[170.255]H | tr B8R341 B8R341 9ARAC | NADH-ubiquinone oxidoreductase chain 1 | Sicarius aff. patagonicus | |
| RT[255.953][231.083][299.111]CVG | tr C1ITN3 C1ITN3 LOXAS | cytochrome c oxidase subunit 1 | Loxosceles aff. spinulosa | |
| I[211.14][268.32]I[187.685]IV[246.077] | tr C1ITP7 C1ITP7 9ARAC | cytochrome c oxidase subunit 1 | Sicarius dolichocephalus | |
| FF[238.18]IITAG[273.235] | tr C1ITR2 C1ITR2 9ARAC | cytochrome c oxidase subunit 1 | Drymusa serrana | |
| YEDRIVVR[230.004] | tr C1ITR3 C1ITR3 9ARAC | cytochrome c oxidase subunit 1 | Drymusa dinora | |
| [316.142]TVYCMSIEITA[226.21]IEDI[241.221] | tr C1IZB2 C1IZB2 LOXLA | cytochrome c oxidase subunit 1 | Loxosceles laeta | |
| GVI[248.157]NITGYR | tr C5J3X9 C5J3X9 LOXRU | cytochrome c oxidase subunit 1 | Loxosceles rufescens | |
| IA[128.08]MFTIIE[216.039]GV[215.166] | tr B8R317 B8R317 LOXAP | NADH-ubiquinone oxidoreductase chain 1 | Loxosceles apachea | |
| [278.338]A[234.386]WHVEN[210.973]VNI[210.062] | tr B8R335 B8R335 LOXS4 | NADH-ubiquinone oxidoreductase chain 1 | Loxosceles sp. | |
| [265.105]V[213.051][172.098]A[283.126]Y[201.071][210.087][278.104][269.011]RGIVMV[283.117] | tr C1ITR3 C1ITR3 9ARAC | cytochrome c oxidase subunit 1 | Drymusa dinora | |
| [127.983]AG[153.948]ACTGEMGSCG | tr C5J3X7 C5J3X7 LOXGA | cytochrome c oxidase subunit 1 | Loxosceles gaucho | |
| YSVFCMM[128.12]V | tr C5J3X9 C5J3X9 LOXRU | cytochrome c oxidase subunit 1 | Loxosceles rufescens | |
| [225.309]SWD[217.039][271.219]C[258.226][218.143][302.82]GMIGSEN | tr Q6W974 Q6W974 LOXRE | sodium/potassium ATPase alpha subunit | Loxosceles reclusa |
Taken together, the identified toxins in the L. intermedia venom include representatives from all toxin groups, even if in low abundances (as in the case of, e.g., hyaluronidases and serine proteases). We also find it noteworthy that we obtained significant coverage of the three major families present in the venom, viz., PLDs, astacin-like metalloproteases, and ICK peptides. These families are of great importance for studies of the brown-spider envenomation features and of biotechnological and medical applications.
Many of the aligned contigs mapped to distinct PLD isoforms from a variety of Loxosceles species. In fact, these toxins are the most studied and well-characterized components of the Loxosceles venom5,20,26,31,46–48. PLDs are able to reproduce the deleterious effects observed in loxoscelism and represent a great target for drug discovery against brown-spider envenomation2,5.
As for the astacin-like metalloproteases identified, we note that astacins were first described as an animal-venom component in 2007 (ref. 28) and only later recognized as a family of toxins present in the Loxosceles venom33. These toxins present proteolytic activity on distinct extracellular matrix proteins and are related to the hemostatic effects in loxoscelism43,49.
ICK peptides, the major components of the L. intermedia venom-gland transcriptome (54,9% of the expressed sequence tags), were identified with correspondence to all four different ICK peptides described for L. intermedia (LiTx1, LiTx2, LiTx3, and LiTx4)50,51. These ICK peptides, also called knottins, are characterized by the neurotoxic properties they exhibit on ion channels and receptors expressed in the nervous systems of insects and mammals52. The high expression of LiTx transcripts, which correlates with the proteomic results found herein, are consistent with the venom’s effects of paralyzing and killing both preys and predators1,20,51.
Additional Information
How to cite this article: Trevisan-Silva, D. et al. A multi-protease, multi-dissociation, bottom-up-to-top-down proteomic view of the Loxosceles intermedia venom. Sci. Data 4:170090 doi: 10.1038/sdata.2017.90 (2017).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors thank CNPq and CAPES for financial support. They also thank Fiocruz for use of Mass Spectrometry Facility RPT02H at the Carlos Chagas Institute, as well as Dr Michel Batista for aiding in the mass spectrometry procedures. V.C.B. acknowledges support from a FAPERJ BBP grant. N.B. is an Alfred P. Sloan Research Fellow and was partially supported by the US National Institutes of Health Grant 2 P41 GM103484-06A1 from the National Institute of General Medical Sciences. The authors thank Wagner Nagib from Carlos Chagas Instituto, Fiocruz—Paraná for generating the final cover art.
Footnotes
N.B. has an equity interest in Digital Proteomics, LLC, a company that may potentially benefit from the research results; Digital Proteomics, LLC was not involved in any aspects of this research. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. The remaining authors declares no competing financial interests.
Data Citations
- Trevisan-Silva D. 2016. Dummy. PXD005523
- Trevisan-Silva D. 2017. Figshare. https://doi.org/10.6084/m9.figshare.c.3709168
References
- Escoubas P. Molecular diversification in spider venoms: a web of combinatorial peptide libraries. Mol. Divers. 10, 545–554 (2006). [DOI] [PubMed] [Google Scholar]
- Gremski L. H. et al. Recent advances in the understanding of brown spider venoms: From the biology of spiders to the molecular mechanisms of toxins. Toxicon Off. J. Int. Soc. Toxinology 83, 91–120 (2014). [DOI] [PubMed] [Google Scholar]
- You K. E. et al. The effective control of a bleeding injury using a medical adhesive containing batroxobin. Biomed. Mater. Bristol Engl 9, 025002 (2014). [DOI] [PubMed] [Google Scholar]
- Byron M., Zochert S., Hellwig T., Gavozdea-Barna M. & Gulseth M. P. Successful use of laboratory monitoring to facilitate an invasive procedure for a patient treated with dabigatran. Am. J. Health-Syst. Pharm. AJHP Off. J. Am. Soc. Health-Syst. Pharm. 74, 461–465 (2017). [DOI] [PubMed] [Google Scholar]
- Chaim O. M. et al. Brown spider (Loxosceles genus) venom toxins: tools for biological purposes. Toxins 3, 309–344 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves-Ferreira A. G. C. et al. Structural and functional analyses of DM43, a snake venom metalloproteinase inhibitor from Didelphis marsupialis serum. J. Biol. Chem. 277, 13129–13137 (2002). [DOI] [PubMed] [Google Scholar]
- Rocha S. L. G. et al. Functional analysis of DM64, an antimyotoxic protein with immunoglobulin-like structure from Didelphis marsupialis serum. Eur. J. Biochem. 269, 6052–6062 (2002). [DOI] [PubMed] [Google Scholar]
- Mendes T. M. et al. Generation and characterization of a recombinant chimeric protein (rCpLi) consisting of B-cell epitopes of a dermonecrotic protein from Loxosceles intermedia spider venom. Vaccine 31, 2749–2755 (2013). [DOI] [PubMed] [Google Scholar]
- Gui J. et al. A tarantula-venom peptide antagonizes the TRPA1 nociceptor ion channel by binding to the S1-S4 gating domain. Curr. Biol. CB 24, 473–483 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung A. R. et al. The effect of PnTx2-6 protein from Phoneutria nigriventer spider toxin on improvement of erectile dysfunction in a rat model of cavernous nerve injury. Urology 84, 730.e9–17 (2014). [DOI] [PubMed] [Google Scholar]
- Liu Z. et al. A novel spider peptide toxin suppresses tumor growth through dual signaling pathways. Curr. Mol. Med. 12, 1350–1360 (2012). [DOI] [PubMed] [Google Scholar]
- Zobel-Thropp P. A., Bodner M. R. & Binford G. J. Comparative analyses of venoms from American and African Sicarius spiders that differ in sphingomyelinase D activity. Toxicon Off. J. Int. Soc. Toxinology 55, 1274–1282 (2010). [DOI] [PubMed] [Google Scholar]
- King G. F. & Hardy M. C. Spider-venom peptides: structure, pharmacology, and potential for control of insect pests. Annu. Rev. Entomol. 58, 475–496 (2013). [DOI] [PubMed] [Google Scholar]
- Junqueira M. & Carvalho P. C. Tools and challenges for diversity-driven proteomics in Brazil. Proteomics 12, 2601–2606 (2012). [DOI] [PubMed] [Google Scholar]
- Eng J. K., McCormack A. L. & Yates J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom 5, 976–989 (1994). [DOI] [PubMed] [Google Scholar]
- Perkins D. N., Pappin D. J., Creasy D. M. & Cottrell J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999). [DOI] [PubMed] [Google Scholar]
- Seidler J., Zinn N., Boehm M. E. & Lehmann W. D. De novo sequencing of peptides by MS/MS. Proteomics 10, 634–649 (2010). [DOI] [PubMed] [Google Scholar]
- Futrell J. M. Loxoscelism. Am. J. Med. Sci. 304, 261–267 (1992). [DOI] [PubMed] [Google Scholar]
- Fernandes-Pedrosa M. de F. et al. Transcriptome analysis of Loxosceles laeta (Araneae, Sicariidae) spider venomous gland using expressed sequence tags. BMC Genomics 9, 279 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremski L. H. et al. A novel expression profile of the Loxosceles intermedia spider venomous gland revealed by transcriptome analysis. Mol. Biosyst. 6, 2403–2416 (2010). [DOI] [PubMed] [Google Scholar]
- Binford G. J. & Wells M. A. The phylogenetic distribution of sphingomyelinase D activity in venoms of Haplogyne spiders. Comp. Biochem. Physiol. B Biochem. Mol. Biol 135, 25–33 (2003). [DOI] [PubMed] [Google Scholar]
- Machado L. F. et al. Proteome analysis of brown spider venom: identification of loxnecrogin isoforms in Loxosceles gaucho venom. Proteomics 5, 2167–2176 (2005). [DOI] [PubMed] [Google Scholar]
- dos Santos L. D., Dias N. B., Roberto J., Pinto A. S. & Palma M. S. Brown recluse spider venom: proteomic analysis and proposal of a putative mechanism of action. Protein Pept. Lett 16, 933–943 (2009). [DOI] [PubMed] [Google Scholar]
- Washburn M. P., Wolters D. & Yates J. R. 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 (2001). [DOI] [PubMed] [Google Scholar]
- Binford G. J., Cordes M. H. J. & Wells M. A. Sphingomyelinase D from venoms of Loxosceles spiders: evolutionary insights from cDNA sequences and gene structure. Toxicon Off. J. Int. Soc. Toxinology 45, 547–560 (2005). [DOI] [PubMed] [Google Scholar]
- Chaim O. M. et al. Brown spider dermonecrotic toxin directly induces nephrotoxicity. Toxicol. Appl. Pharmacol. 211, 64–77 (2006). [DOI] [PubMed] [Google Scholar]
- da Silveira R. B. et al. Molecular cloning and functional characterization of two isoforms of dermonecrotic toxin from Loxosceles intermedia (brown spider) venom gland. Biochimie 88, 1241–1253 (2006). [DOI] [PubMed] [Google Scholar]
- da Silveira R. B. et al. Identification, cloning, expression and functional characterization of an astacin-like metalloprotease toxin from Loxosceles intermedia (brown spider) venom. Biochem. J. 406, 355–363 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silveira R. B. et al. Hyaluronidases in Loxosceles intermedia (Brown spider) venom are endo-beta-N-acetyl-d-hexosaminidases hydrolases. Toxicon Off. J. Int. Soc. Toxinology 49, 758–768 (2007). [DOI] [PubMed] [Google Scholar]
- da Silveira R. B. et al. Two novel dermonecrotic toxins LiRecDT4 and LiRecDT5 from brown spider (Loxosceles intermedia) venom: from cloning to functional characterization. Biochimie 89, 289–300 (2007). [DOI] [PubMed] [Google Scholar]
- Appel M. H. et al. Identification, cloning and functional characterization of a novel dermonecrotic toxin (phospholipase D) from brown spider (Loxosceles intermedia) venom. Biochim. Biophys. Acta 1780, 167–178 (2008). [DOI] [PubMed] [Google Scholar]
- de Santi Ferrara G. I. et al. SMase II, a new sphingomyelinase D from Loxosceles laeta venom gland: molecular cloning, expression, function and structural analysis. Toxicon Off. J. Int. Soc. Toxinology 53, 743–753 (2009). [DOI] [PubMed] [Google Scholar]
- Trevisan-Silva D. et al. Differential metalloprotease content and activity of three Loxosceles spider venoms revealed using two-dimensional electrophoresis approaches. Toxicon Off. J. Int. Soc. Toxinology 76, 11–22 (2013). [DOI] [PubMed] [Google Scholar]
- Trevisan-Silva D. et al. Astacin-like metalloproteases are a gene family of toxins present in the venom of different species of the brown spider (genus Loxosceles). Biochimie 92, 21–32 (2010). [DOI] [PubMed] [Google Scholar]
- Jeong K., Kim S. & Pevzner P. A. UniNovo: a universal tool for de novo peptide sequencing. Bioinformatics 29, 1953–1962 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthals A., Clauser K. R., Frank A. M. & Bandeira N. Sequencing-Grade De novo Analysis of MS/MS Triplets (CID/HCD/ETD) From Overlapping Peptides. J. Proteome Res. 12, 2846–2857 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthals A., Clauser K. R. & Bandeira N. Shotgun protein sequencing with meta-contig assembly. Mol. Cell. Proteomics MCP 11, 1084–1096 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandeira N. Spectral networks: a new approach to de novo discovery of protein sequences and posttranslational modifications. BioTechniques, 691 passim 42, 687 (2007). [DOI] [PubMed] [Google Scholar]
- Leprevost F. V. et al. PepExplorer: A Similarity-driven Tool for Analyzing de Novo Sequencing Results. Mol. Cell. Proteomics MCP 13, 2480–2489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catherman A. D., Skinner O. S. & Kelleher N. L. Top Down proteomics: Facts and perspectives. Biochem. Biophys. Res. Commun. 445, 683–693 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M. L. & Vasconcellos-Neto J. Microhabitats occupied by Loxosceles intermedia and Loxosceles laeta (Araneae: Sicariidae) in Curitiba, Paraná, Brazil. J. Med. Entomol. 42, 756–765 (2005). [DOI] [PubMed] [Google Scholar]
- Choudhary G., Wu S.-L., Shieh P. & Hancock W. S. Multiple enzymatic digestion for enhanced sequence coverage of proteins in complex proteomic mixtures using capillary LC with ion trap MS/MS. J. Proteome Res. 2, 59–67 (2003). [DOI] [PubMed] [Google Scholar]
- Feitosa L. et al. Detection and characterization of metalloproteinases with gelatinolytic, fibronectinolytic and fibrinogenolytic activities in brown spider (Loxosceles intermedia) venom. Toxicon Off. J. Int. Soc. Toxinology 36, 1039–1051 (1998). [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- Vizcaíno J. A. et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res 41, D1063–D1069 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalapothakis E. et al. Molecular cloning, expression and immunological properties of LiD1, a protein from the dermonecrotic family of Loxosceles intermedia spider venom. Toxicon Off. J. Int. Soc. Toxinology 40, 1691–1699 (2002). [DOI] [PubMed] [Google Scholar]
- Ribeiro R. O. S. et al. Biological and structural comparison of recombinant phospholipase D toxins from Loxosceles intermedia (brown spider) venom. Toxicon Off. J. Int. Soc. Toxinology 50, 1162–1174 (2007). [DOI] [PubMed] [Google Scholar]
- Vuitika L. et al. Brown spider phospholipase-D containing a conservative mutation (D233E) in the catalytic site: identification and functional characterization. J. Cell. Biochem. 114, 2479–2492 (2013). [DOI] [PubMed] [Google Scholar]
- da Silveira R. B. et al. Identification of proteases in the extract of venom glands from brown spiders. Toxicon Off. J. Int. Soc. Toxinology 40, 815–822 (2002). [DOI] [PubMed] [Google Scholar]
- de Castro C. S. et al. Identification and molecular cloning of insecticidal toxins from the venom of the brown spider Loxosceles intermedia. Toxicon Off. J. Int. Soc. Toxinology 44, 273–280 (2004). [DOI] [PubMed] [Google Scholar]
- Matsubara F. H. et al. A novel ICK peptide from the Loxosceles intermedia (brown spider) venom gland: cloning, heterologous expression and immunological cross-reactivity approaches. Toxicon Off. J. Int. Soc. Toxinology 71, 147–158 (2013). [DOI] [PubMed] [Google Scholar]
- Dutertre S. & Lewis R. J. Use of venom peptides to probe ion channel structure and function. J. Biol. Chem. 285, 13315–13320 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Chambers M. C. & Tabb D. L. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J. Proteome Res. 6, 3549–3557 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Trevisan-Silva D. 2016. Dummy. PXD005523
- Trevisan-Silva D. 2017. Figshare. https://doi.org/10.6084/m9.figshare.c.3709168