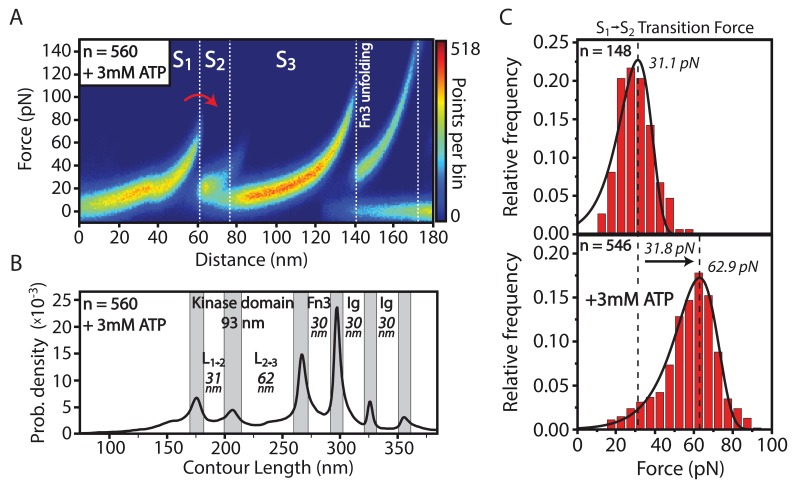
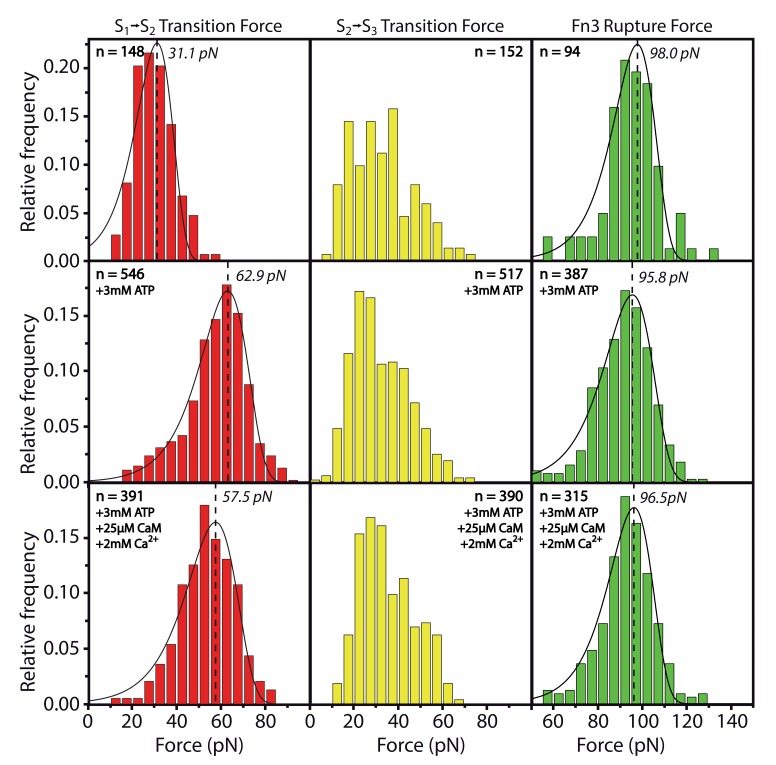
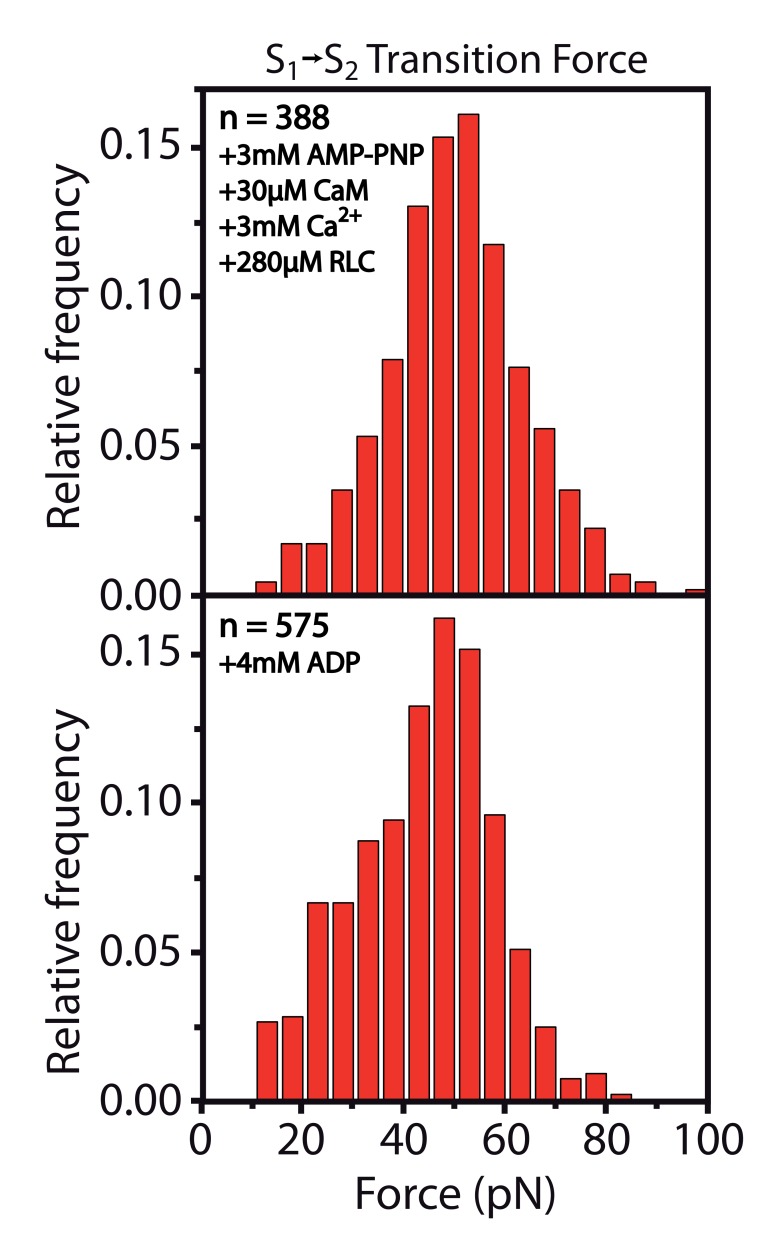
Figure 2. Structural effects of ATP binding on smMLCK’s characteristic sequence of conformational states.
(A) Stabilization of the S→S transition upon ligand binding. For better illustration, a heatmap of 560 aligned curves is depicted. (B) Contour-length transformation of the respective events in the presence of ATP. L is associated to the contour length released at the transition from state S to S. (C) Statistical evaluation of S→S stabilization via force histograms fitted with the Bell-Evans model. An increase in the most probable transition force of about 30 pN is observed upon ATP addition. Both data sets were recorded within one experiment with the same cantilever.
DOI: http://dx.doi.org/10.7554/eLife.26473.004