Figure 2.
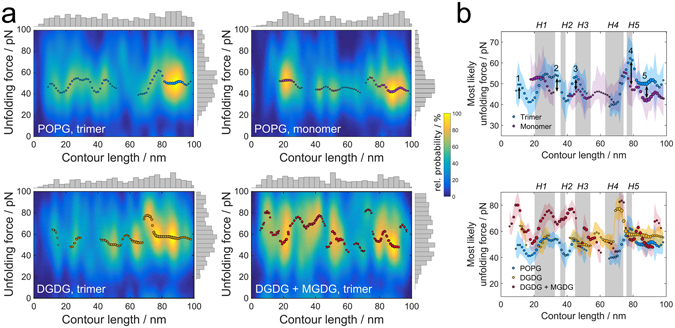
Unfolding forces of LHCII depend on trimerization and the lipid matrix. (a) Multivariate kernel density estimate of the unfolding force as a function of the contour length for LHCII monomers in POPG (top right) and LHCII trimers in POPG (top left), pure DGDG (bottom left) and a mixture of DGDG and MGDG (bottom right). The dots mark the most likely unfolding forces for a given contour length, while the diameter of the dots represents their relative probability of at least 40% in relation to the most probable unfolding event. The histograms (grey) at the axes show the individual underlying distributions of unfolding forces and contour lengths. (b) Most likely unfolding force as a function of the contour length (obtained from the dots in (a)) as a function of oligomerization state (top) and lipid mixture (bottom). Helices (H1-5) are indicated as shaded gray areas. The color shaded areas indicate the standard error of the most likely unfolding force. The trimer shows enhanced stabilization compared to the monomer in distinct domains of the protein, indicated by numbers 1 to 5. Both glycolipids provide additional stability for trimeric LHCII at helix 4, while the addition of MGDG strongly increases the unfolding forces in many segments along the LHCII polypeptide.
