Figure 3.
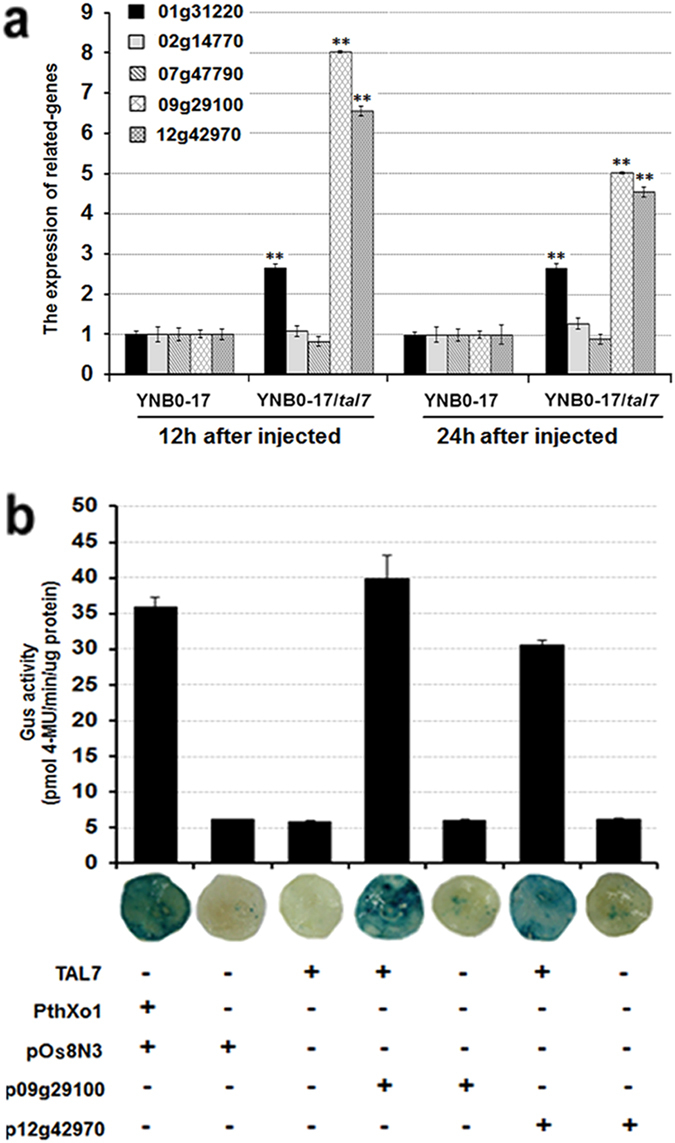
Xoc Tal7 targets rice genes Os09g29100 and Os12g42970. (a) The expression of candidate targets of Tal7. Xoc strains YNB0-17, and YNB0-17(tal7) were infiltrated into IR24 rice, and the expression of five rice genes (Os01g31220, Os02g14770, Os07g47790, Os09g29100, and Os12g42970) was measured 12 and 24 hpi by real time qRT-PCR. The expression levels of Actin and 18S rRNA used as internal standards. The asterisks in each horizontal data column indicate significant differences at P = 0.01 using the Student’s t test. Data are the mean ± SD of triplicate measurements from a representative experiment; and similar results were obtained in two other independent experiments. (b) Transcriptional activation of rice genes Os09g29100 and Os12g42970 by TALEs of Xoc Tal7. The TALE PthXo1 from Xoo and its target rice gene, Os8N3 (pOs8N3), were used as a positive control. Reporter fusions containing the rice promoters fused to GUS were codelivered via A. tumefaciens into N. benthamiana with (+) and without (−) constructs encoding TAL7 and PthXo1. pOs8N3, p09G29100 and p12g42970 represent the Os8N3, 09g29100 and 12g42970 promoters fused to GUS (see Methods). For quantitative assays, two leaf discs (0.9 cm diameter) were sampled 2 dpi, and GUS activity was determined using 4-methyl-umbelliferyl-β-D-glucuronide (MUG). 4-MU is 4-methyl-umbelliferone. Error bars indicate standard deviations (n = 3 samples from different plants). For qualitative assays, GUS activity in excised leaf discs was determined 3 dpi with X-Gluc (5-bromo-4-chloro-3-indolyl-b-D-glucuronide). A blue color indicatesa positive reaction. All experiments were performed twice with similar results.
