Figure 5.
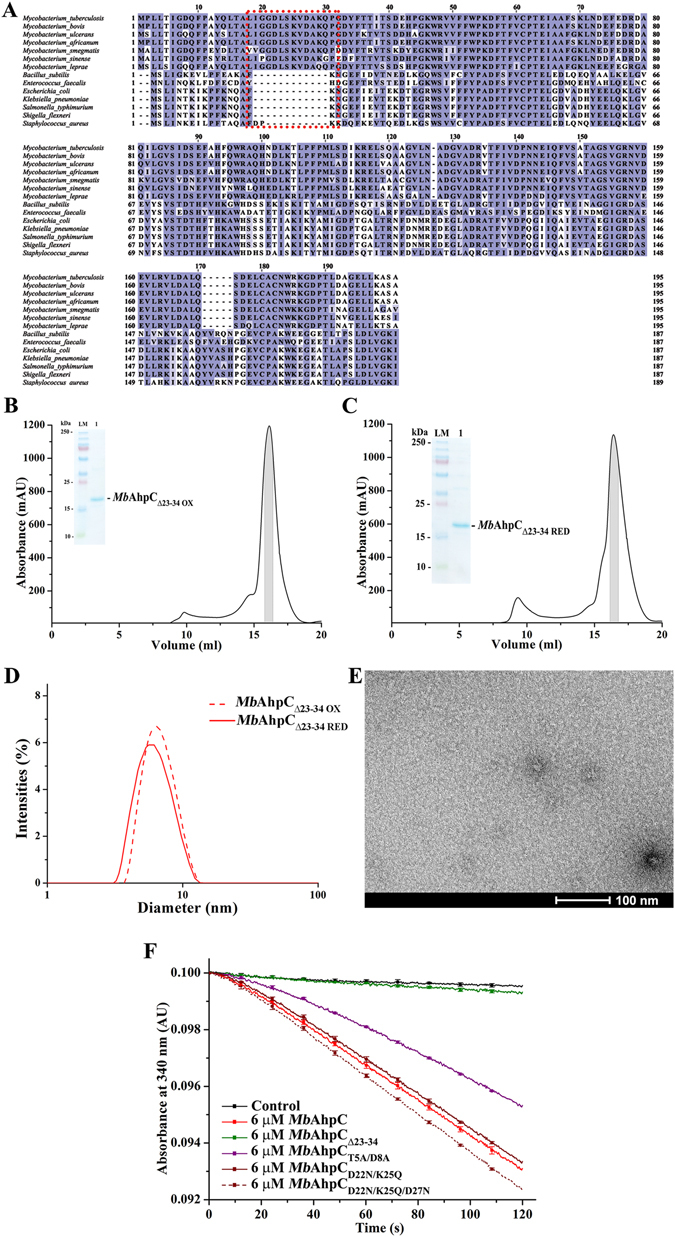
(A) Multiple sequence alignments of AhpC from different bacteria using ClustalW17. As shown, various Mycobacterium sp. presented extra 15 residues located at the N-terminus (red dashed box), which were absent in other bacteria species. Mutations with deletion from residues 23–34 were constructed and both oxidized (B) and reduced (C) mutants eluted at approximately 16 ml on a Superdex 200 HR 10/30 column and showed high purity on a 17% SDS gel (inset). (D) DLS analysis of oxidized (red dashed line) and reduced (red solid line) of MbAhpCΔ23–34. Both redox states of the protein revealed a similar diameter unlike that of wt MbAhpC. (E) Negative stained EM-images of reduced MbAhpCΔ23–34. The lack of ring-shaped particles in the micrograph indicates the inability of MbAhpCΔ23–34 to form an oligomeric ring. (F) NADPH-oxidation of MbAhpC, -AhpCΔ23–34, -AhpCT5A/D8A, -AhpCD22N/K25Q or -AhpCD22N/K25Q/D27N measured at 50 µM H2O2 is shown as a representative to highlight the effects of the mutations generated. A control without MbAhpC is represented with a black solid line. The activity of wt MbAhpC is indicated with a red solid line. The deletion of residues 23–34 (green solid line) altered the activity of the protein, hence, showing activities close to that of which resembles the background activity. However, the mutations in T5 and D8 showed the decline of activity of MbAhpCT5A/D8A as shown with the purple solid line. On the other hand, the enzymatic activity of MbAhpCD22N/K25Q (brown solid line) or MbAhpCD22N/K25Q/D27N (brown dashed line) have similar activity to wt MbAhpC.
