Figure 1. Comparative analysis of SERCA‒SLN and SERCA‒PLB crystal structures.
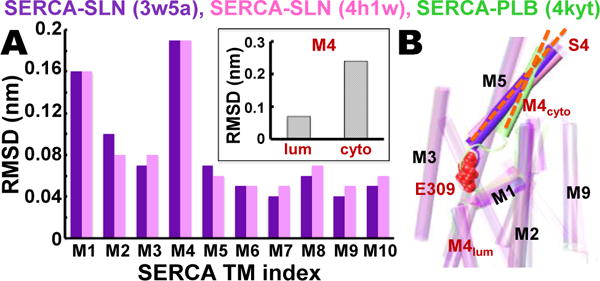
(A) RMSD for each TM helix in SERCA‒SLN crystal structures 3w5a6 (purple) and 4h1w7 (pink) was calculated relative to TM helices in the SERCA‒PLB crystal structure 4kyt22. The inset shows RMSD of M4S4 divided into luminal and cytosolic segments R290-A306 (lum) and P312-K329 (cyto). (B) Superposed crystal structures of SERCA‒SLN (purple, pink) and SERCA‒PLB (green) reveal that SLN, but not PLB, mediates a large increase in the axial tilt angle (orange dashes) of M4S4. SERCA residue E309 (red spacefill) is the Ca2+ binding residue in transport site II in the TM domain that transmits the ‘full-occupancy’ signal to initiate ATP hydrolysis and phosphoenzyme formation at D351 in the phosphorylation domain.23,24
