Fig. 2.
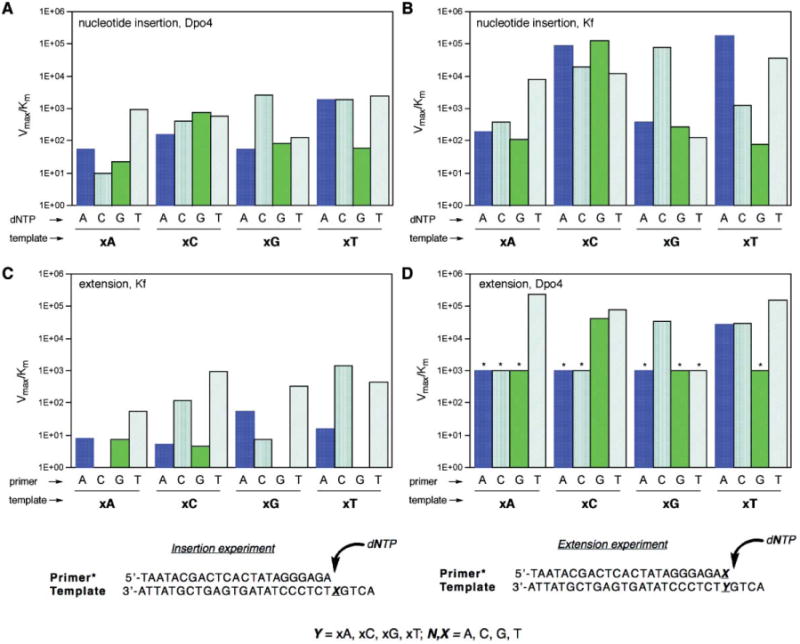
Plots (top) comparing kinetic efficiencies and selectivities for xDNA pair synthesis and extension for Dpo4 and Kf exo- enzymes and primer: templates (bottom) used in single nucleotide insertion and extension experiments. (A) Nucleotide insertion by Dpo4; (B) nucleotide insertion by Kf exo- (data from ref. 13); (C) primer/template base pair extension by Kf exo- (extending various pairs/mismatches shown); (D) base pair extension by Dpo4 (* = estimated maximum value; see Table S3†). In all graphs, units on Vmax/Km = min−1·μM−1.
