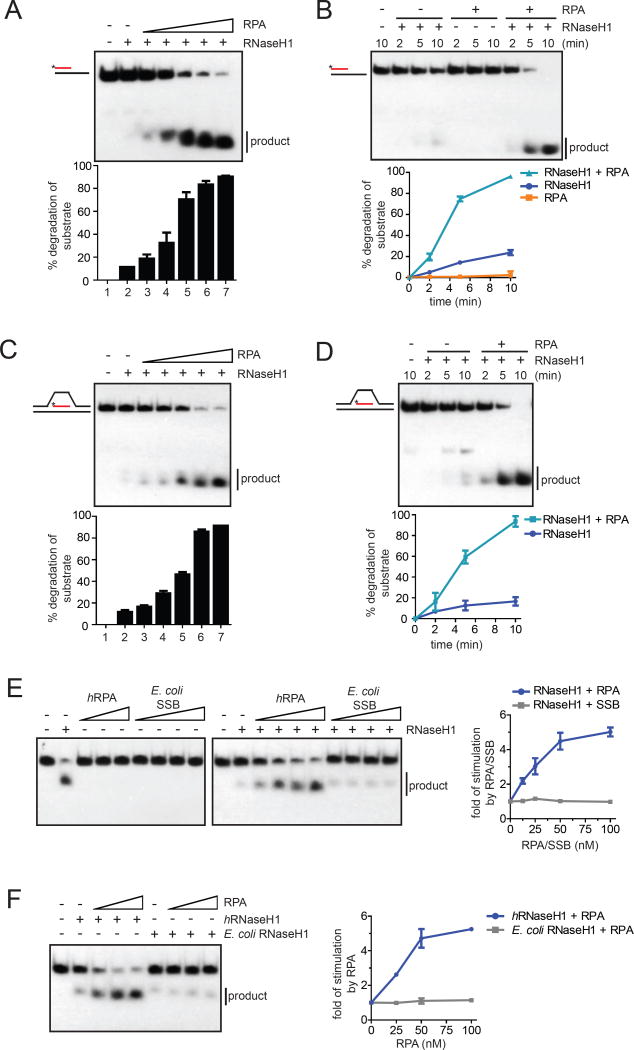
Fig. 2. RPA stimulates the activity of RNaseH1 on R loops.
(A) The R:D+ssDNA substrate with 32P-labeled RNA (25 nM) was incubated with RNaseH1 (2 nM) and increasing concentrations of RPA (0, 12.5, 25, 50, 100, 200 nM) for 5 min. The fractions of substrate cleaved by RNaseH1 were quantified. Data are presented as mean ± SD (n=3). (B) The R:D+ssDNA substrate (100 nM) was incubated with RNaseH1 (5 nM), RPA (100 nM), or both for the indicated amounts of time. Data are presented as mean ± SD (n=3). (C-D) In C, the R loop substrate (25 nM) was incubated with RNaseH1 (2 nM) and increasing concentrations of RPA (0, 12.5, 25, 50, 100, 200 nM) for 5 min. In D, the R loop substrate (100 nM) was incubated with RNaseH1 (2 nM) in the presence or absence of RPA (100 nM) for the indicated amounts of time. Data are presented as mean ± SD (n=3). (E) Left panel, RNA:DNA hybrid (25 nM) was incubated human RPA (0, 12.5*, 25, 50, 100 nM) or E. coli SSB (0, 12.5, 25, 50, 100 nM) in the presence or absence of human RNaseH1 for 5 min. *: only used in the presence of RNaseH1. Right panel, the cleavage of substrate was quantified, and the fold of simulation by RPA was determined. Data are presented as mean ± SD (n=3). (F) Left panel, RNA:DNA hybrid (25 nM) was incubated with human (2 nM) or E. coli RNaseH1 (0.05 nM) and increasing concentrations of human RPA (0, 25, 50, 100 nM). Right panel, the fold of stimulation by RPA was measured as in E. Data are presented as mean ± SD (n=3). See also Fig. S2.