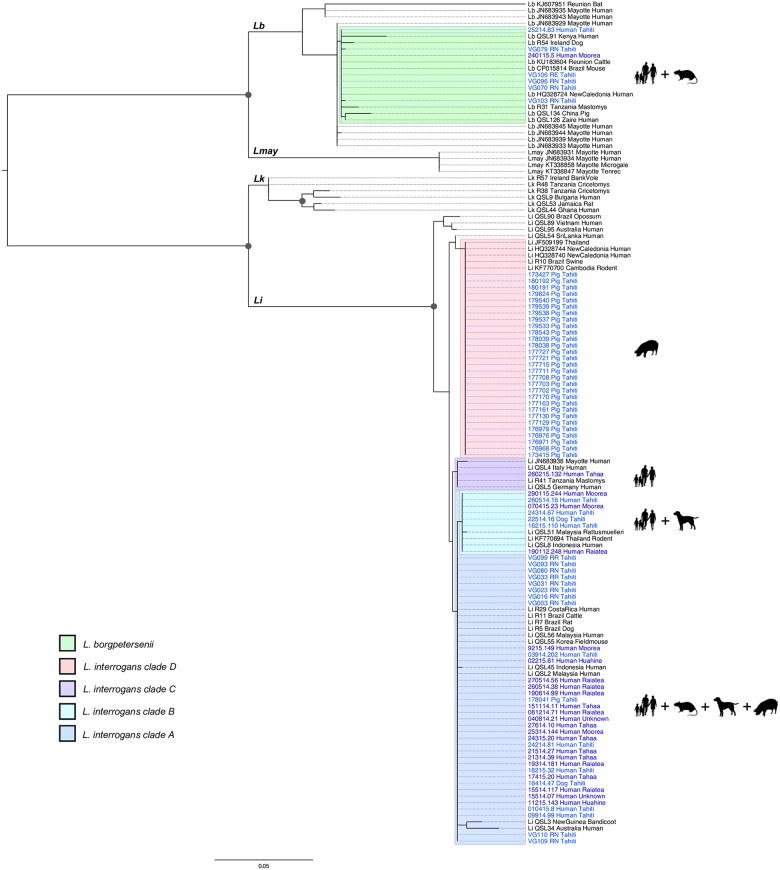
Fig 4. Maximum-likelihood phylogenetic tree (model TN93+G; 1,000 replicates) inferred from secY gene (444-bp sequence).
Human and animal samples from Tahiti are shown in light blue and clinical samples from other islands in French Polynesia (FP) in dark blue; they are referred to using identifiers accompanied by the host name. Corresponding GenBank accession numbers are reported on S6 Table. Published sequences included in the phylogeny are written in black using GenBank accession numbers or ID (for samples from [33]) followed by the country of origin and the host species. Leptospira species are indicated for published sequences: Li: Leptospira interrogans; Lb: Leptospira borgpetersenii; Lk: Leptospira kirschneri; Lmay: Leptospira mayottensis. Bootstrap values higher than 70% are indicated by a dark circle. Black silhouettes represent host groups from FP (i.e. human, dog, pig or rat). Legend refers to the different clusters including samples from FP, with four clades (A to D) inside the species Leptospira interrogans.