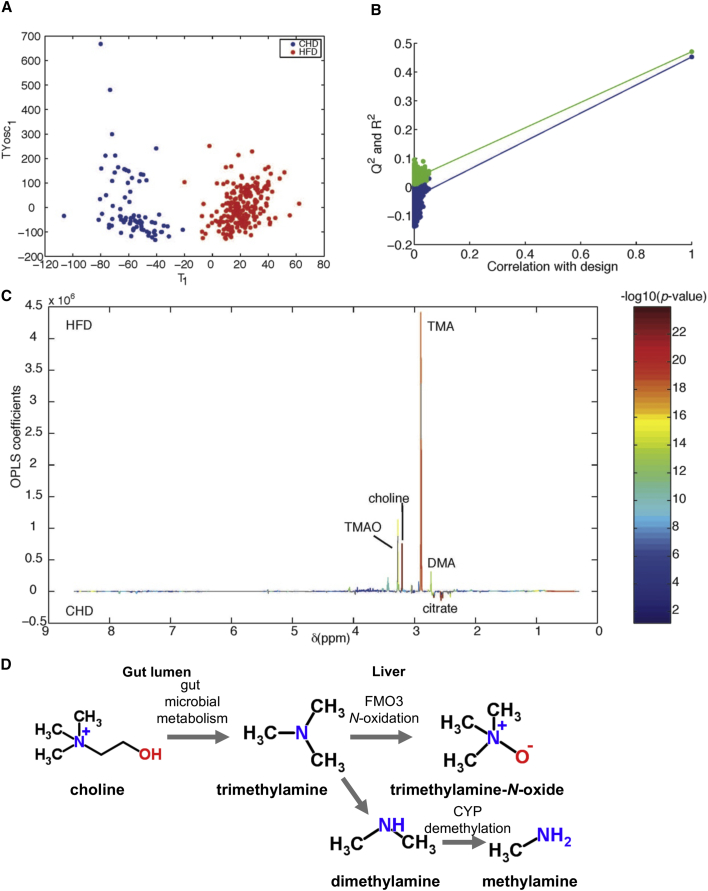
Figure 2.
The Urinary Metabolic Signature of HFD in the C57 Mouse
(A) O-PLS-DA scores plot.
(B) O-PLS-DA permutation plot. The O-PLS-DA model was validated by random permutations (n = 10,000 iterations) of the original variable to explain class membership (CHD versus HFD). The horizontal axis represents the correlation between the original class membership (right) and the randomly permuted class membership vectors (no longer correlated with the original class membership) (left). The y axis represents the goodness-of-fit R2 (in green) parameter obtained for each O-PLS-DA model and the goodness-of-prediction Q2 (in blue) parameter obtained by 7-fold cross-validation of the O-PLS-DA model. The R2 and Q2 parameters for the original model in the top right corner do not belong to the population of 10,000 models fitted with random class memberships, highlighting that the original model does not belong to the population of 10,000 randomly permuted models (p < 0.0001) and thereby confirming the significance of the fitness and prediction ability attached with the original O-PLS-DA model.
(C) O-PLS-DA model coefficient plot.
(D) Summary of microbial-mammalian co-metabolism of methylamines.