Figure 1.
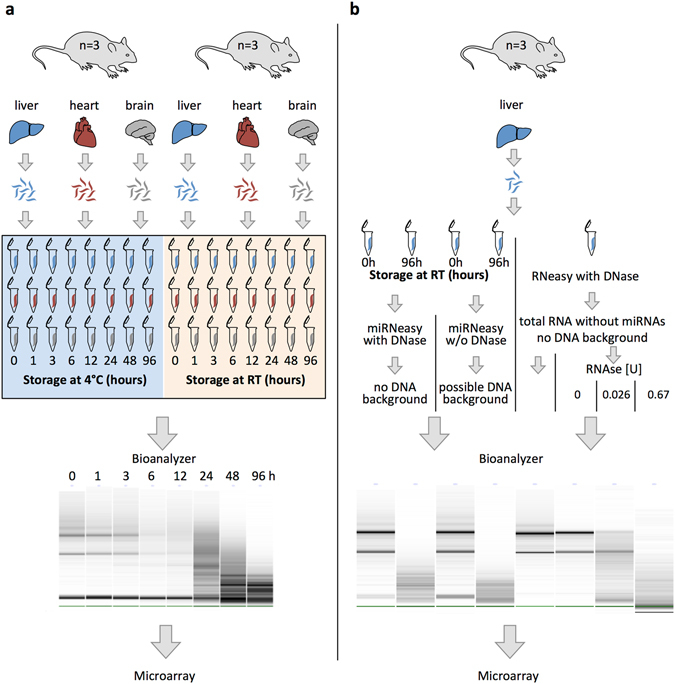
Experimental design. (a) Liver, heart and brain of male mice were harvested immediately after death, divided into 8 parts of about equal size, and stored at either 4 °C or at room temperature (RT) for the indicated time periods before RNA isolation. Experiments were performed in biological triplicates. RNA integrity was measured with Bioanalyzer. Gel-like image of brain tissue is given as example. MiRNA expression profiles of one replicate were measured using microarrays. (b) Liver tissue of 3 male mice was harvested immediately after death and divided into 5 parts of about equal size. Three parts were immediately transferred into RNAlater (0 h), two parts were stored for 96 h at room temperature (96 h). Two samples (0 h and 96 h) were isolated using standard procedure with miRNeasy Kit without DNase digestion. Two samples (0 h and 96 h) were isolated with optional DNase digestion to exclude DNA background. From the remaining undegraded sample (0 h), total RNA without small RNAs was isolated using RNeasy Kit with optional DNase digestion. Isolated RNA was further treated with 0 U, 0.026 U and 0.67 U RNase for 30 min to generate artificial RNA degradation. RNA integrity was measured with Bioanalyzer. MiRNA expression profiles of all replicates were measured using microarrays. The schematic drawings were prepared using the Biomedical-PPT-Toolkit-Suite from Motifolio Inc., USA.
