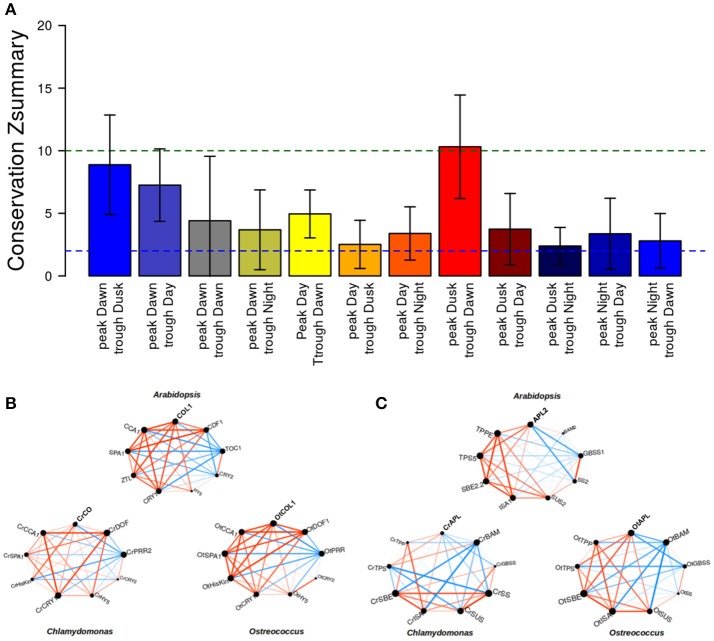
Figure 10.
Conservation of circadian patterns in Arabidopsis, Chlamydomonas, and Ostreococcus. (A) The average and standard deviation of the Zsummary conservation score between the three different species was computed for all clusters capturing different daily rhythmic patterns. Only clusters that exhibit a periodic pattern with peak or trough at Dusk or Dawn are significantly conserved over the three different species. (B) The co-expression pattern of the key circadian/photoperiod regulators exhibits a high level of conservation in the three species, but it is more evident for Arabidopsis and Ostreococcus. Red edges represent positive correlation between the corresponding gene expression profiles whereas blue edges indicate negative correlation. Edge width is proportional to the absolute value of the correlation between the corresponding expression profiles. (C) The co-expression patterns between key enzymes in starch/sucrose metabolism are highly conserved among the three different species with slight differences in Chlamydomonas.