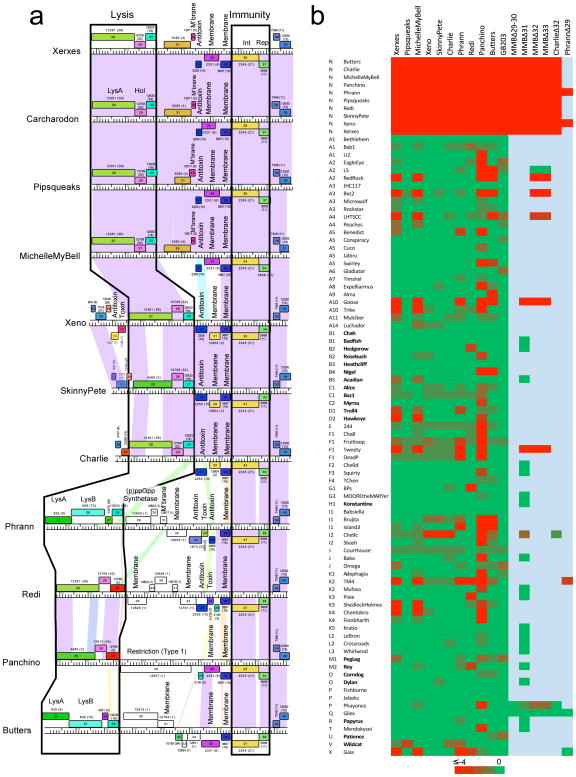
Figure 4. Cluster N prophage-mediated defense against phage infection.
(a) The central parts of 11 Cluster N mycobacteriophage genomes are aligned by their immunity cassettes and putative gene functions are indicated. Genomes are displayed as described for Figure 2, but ordered such that genomes with similarities in these regions are adjacent to each other, particularly Xerxes, Carcharodon, Pipsqueaks, and MMB as one group, and Xeno, SkinnyPete and Charlie as a second group. Note that in Pipsqueaks the open reading frame corresponding to MMB 29 is interrupted by a 25 bp deletion, leaving only the 3’ end of the gene intact and presumably inactivating it. (b) Heat map of prophage-mediated viral sensitivities, reporting efficiencies of plating of mycobacteriophages on Cluster N lysogenic strains or their deletion derivatives. Individual genome names and cluster designations are shown on the left. Phages that do not have an integrase gene and are presumably lytic are shown in bold type; all others are temperate or derivatives of temperate phages. Bright green and bright red corresponds to efficiencies of platings of 1 and less than 10−4, respectively. All efficiencies of plating were determined in at least two separate experiments. Efficiencies of platings are reported in Supplementary Table 2.